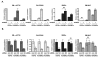NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1 - PubMed (original) (raw)
NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1
Hua Liu et al. Circ Res. 2009.
Abstract
Endothelial cells and mural cells (smooth muscle cells, pericytes, or fibroblasts) are known to communicate with one another. Their interactions not only serve to support fully functional blood vessels but also can regulate vessel assembly and differentiation or maturation. In an effort to better understand the molecular components of this heterotypic interaction, we used a 3D model of angiogenesis and screened for genes, which were modulated by coculturing of these 2 different cell types. In doing so, we discovered that NOTCH3 is one gene whose expression is robustly induced in mural cells by coculturing with endothelial cells. Knockdown by small interfering RNA revealed that NOTCH3 is necessary for endothelial-dependent mural cell differentiation, whereas overexpression of NOTCH3 is sufficient to promote smooth muscle gene expression. Moreover, NOTCH3 contributes to the proangiogenic abilities of mural cells cocultured with endothelial cells. Interestingly, we found that the expression of NOTCH3 is dependent on Notch signaling, because the gamma-secretase inhibitor DAPT blocked its upregulation. Furthermore, in mural cells, a dominant-negative Mastermind-like1 construct inhibited NOTCH3 expression, and endothelial-expressed JAGGED1 was required for its induction. Additionally, we demonstrated that NOTCH3 could promote its own expression and that of JAGGED1 in mural cells. Taken together, these data provide a mechanism by which endothelial cells induce the differentiation of mural cells through activation and induction of NOTCH3. These findings also suggest that NOTCH3 has the capacity to maintain a differentiated phenotype through a positive-feedback loop that includes both autoregulation and JAGGED1 expression.
Figures
Figure 1. Endothelial cells and mural cells interact
Images of blood vessels formed by endothelial cells (HUVECs) cultured alone (A), or cocultured with human dermal neonatal fibroblasts (HDFNs) (B), human umbilical artery smooth muscle cells (HUASMCs) (C) and human coronary artery smooth muscle cells (HCASMCs) (D) in a 3-dimensional angiogenesis assay. Cells were grown in a collagen matrix for five days, fixed, and stained with TRITC labeled endothelial-specific lectin (red), 100× magnification. (E) Triple labeling with lectin (red), a preloaded CellTracker dye (green) to visualize HDFNs surrounding vessels, and DAPI (blue), 400× magnification.
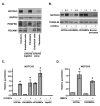
Figure 2. Endothelial cells induce NOTCH3 expression in mural cells
(A) Endothelial cells (HUVECs) and fibroblasts (HDFNs) were cultured alone or cocultured in 3-dimensional angiogenesis assays. After five days, cells were separated by anti-PECAM1-conjugated Dynabeads for Western blot analysis. The purity of the fibroblast and endothelial fractions were evaluated by probing for PDGFRβ as a marker for fibroblasts and PECAM1 as a marker for endothelial cells. (B) HDFNs, HUASMCs, HCASMCs, and bovine pericytes were cultured in 2-dimensions in the presence or absence of HUVECs for 48 hours, separated and subjected to Western analysis. Numbers reflect relative protein expression determined by average pixel intensity from 3 experiments normalized to respective control, P < 0.05. (C) qPCR analysis of NOTCH3 mRNA in HDFNs, HUASMCs, HCASMCs, and HepG2, cultured with or without HUVECs. (D) qPCR analysis of NOTCH3 mRNA in HDFNs and HUASMCs which were cultured alone or cocultured with HMECs. * P < 0.05 compared to control.
Figure 3. Notch signaling is activated in mural cells upon coculture with endothelial cells
(A) Luciferase reporter assay to assess Notch activity. HDFNs were transfected with pGL3-promoter-luciferase (pGL3) plasmid as control, or plasmid with 5 CBF1 binding elements upstream of the promoter (5XCBF1), cultured in the presence or absence of HUVECs, and luciferase activity was measured. RLU, relative light units. (B, C) qPCR analysis of Notch target genes, HEYL/HRT3 and HES1 in mural cells cultured with or without HUVECs. (D) Knockdown of NOTCH3 by siRNA in HDFNs, HUASMCs and HCASMCs was confirmed by qPCR and (E) Western blot analysis. (F) qPCR analysis of HEYL/HRT3 mRNA expression after NOTCH3 knockdown. Mural cells were transiently transfected with control siRNA (con) or NOTCH3 siRNA (N3). HUVECs were added, and cocultured for an additional 48 hours. * P < 0.05 relative to relevant control; ns, not significant.
Figure 4. Endothelial-induced smooth muscle gene expression is dependent on NOTCH3
(A) Fibroblasts and smooth muscle cells were cultured with or without HUVECs, separated by anti-PECAM1-conjugated Dynabeads and RNA was collected for qPCR to detect SM α–ACTIN, CALPONIN, SM22α, and SM-MHC transcripts. (B) HDFNs, HUASMCs and HCASMCs were transfected with control siRNA (con) or NOTCH3 siRNA (N3), and cocultured with HUVECs as described. RNA was extracted for qPCR. Significant differences (P < 0.05) between control and respective experimental values were observed in all but those noted as not significant (ns).
Figure 5. Endothelial-induced NOTCH3 expression requires Notch signaling
(A) HUVECs and HDFNs were plated on either side of the transwell insert as indicated, cultured for 48 hours, and the cells from the top surface were harvested for qPCR to examine NOTCH3 mRNA expression. (B) Western blot to detect NOTCH3 expression in HDFNs cocultured with HUVECs with varied amounts of the γ-secretase inhibitor (DAPT). Fibroblasts cultured alone were used as a control. (C) qPCR to measure NOTCH3 mRNA expression in HDFNs cocultured with HUVECs in the presence or absence of DAPT. (D-F) HDFNs were lentivirally transduced with GFP or DN-MAML, then cultured alone or cocultured with HUVECs. Fibroblasts were separated from HUVECs for Western analysis to detect NOTCH3 protein (D), or for qPCR to measure NOTCH3 (E) and HEYL/HRT3 (F) mRNA. * P < 0.05; ns, not significant.
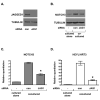
Figure 6. JAGGED1 on endothelial cells is necessary for NOTCH3 induction
HUVECs were transiently transfected with control siRNA (con) or JAGGED1-specific siRNA (JAG1). HDFNs were added, cocultured for 48 hours, and the cells separated by Dynabeads for analysis. (A) Western blot to confirm knockdown of JAGGED1 protein in HUVECs. (B) Western blot for NOTCH3 protein expression. (C, D) qPCR to detect NOTCH3 and HEYL/HRT3 mRNA. * P < 0.05 relative to respective control.
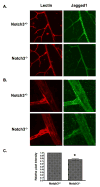
Figure 7. Jagged1 expression is reduced in the absence of Notch3 in vivo
Retinas isolated from Notch3 null (Notch-/-) and heterozygous (Notch+/-) mice at postnatal day 15 were immunostained to detect Jagged1 protein (green). An endothelial-specific lectin (red) was used to highlight the vasculature. Confocal images were taken at (A) 400× and (B) 630× (zoom2) magnification and pixel intensity of Jagged1 staining was quantified (C). Graph represents average Jagged1 expression from 8 null and 8 heterozygous retinas. * P = 0.0004.
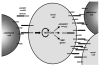
Figure 8. Model of sustained Notch signaling in mural cells
Comment in
- Building a vessel wall with notch signaling.
Regan JN, Majesky MW. Regan JN, et al. Circ Res. 2009 Feb 27;104(4):419-21. doi: 10.1161/CIRCRESAHA.109.194233. Circ Res. 2009. PMID: 19246684 Free PMC article. No abstract available.
Similar articles
- Endothelial cells downregulate apolipoprotein D expression in mural cells through paracrine secretion and Notch signaling.
Pajaniappan M, Glober NK, Kennard S, Liu H, Zhao N, Lilly B. Pajaniappan M, et al. Am J Physiol Heart Circ Physiol. 2011 Sep;301(3):H784-93. doi: 10.1152/ajpheart.00116.2011. Epub 2011 Jun 24. Am J Physiol Heart Circ Physiol. 2011. PMID: 21705670 Free PMC article. - Regulation of vascular smooth muscle cell phenotype in three-dimensional coculture system by Jagged1-selective Notch3 signaling.
Bhattacharyya A, Lin S, Sandig M, Mequanint K. Bhattacharyya A, et al. Tissue Eng Part A. 2014 Apr;20(7-8):1175-87. doi: 10.1089/ten.TEA.2013.0268. Epub 2014 Feb 10. Tissue Eng Part A. 2014. PMID: 24138322 Free PMC article. - Building a vessel wall with notch signaling.
Regan JN, Majesky MW. Regan JN, et al. Circ Res. 2009 Feb 27;104(4):419-21. doi: 10.1161/CIRCRESAHA.109.194233. Circ Res. 2009. PMID: 19246684 Free PMC article. No abstract available. - Blockade of Jagged/Notch pathway abrogates transforming growth factor β2-induced epithelial-mesenchymal transition in human retinal pigment epithelium cells.
Chen X, Xiao W, Liu X, Zeng M, Luo L, Wu M, Ye S, Liu Y. Chen X, et al. Curr Mol Med. 2014 May;14(4):523-34. doi: 10.2174/1566524014666140331230411. Curr Mol Med. 2014. PMID: 24694299 Review. - New models to study vascular mural cell embryonic origin: implications in vascular diseases.
Sinha S, Santoro MM. Sinha S, et al. Cardiovasc Res. 2018 Mar 15;114(4):481-491. doi: 10.1093/cvr/cvy005. Cardiovasc Res. 2018. PMID: 29385541 Review.
Cited by
- Tumor Vascular Morphology Undergoes Dramatic Changes during Outgrowth of B16 Melanoma While Proangiogenic Gene Expression Remains Unchanged.
Langenkamp E, Vom Hagen FM, Zwiers PJ, Moorlag HE, Schouten JP, Hammes HP, Gouw AS, Molema G. Langenkamp E, et al. ISRN Oncol. 2011;2011:409308. doi: 10.5402/2011/409308. Epub 2011 Dec 4. ISRN Oncol. 2011. PMID: 22235379 Free PMC article. - Signaling required for blood vessel maintenance: molecular basis and pathological manifestations.
Murakami M. Murakami M. Int J Vasc Med. 2012;2012:293641. doi: 10.1155/2012/293641. Epub 2011 Dec 6. Int J Vasc Med. 2012. PMID: 22187650 Free PMC article. - Activation dynamics and signaling properties of Notch3 receptor in the developing pulmonary artery.
Ghosh S, Paez-Cortez JR, Boppidi K, Vasconcelos M, Roy M, Cardoso W, Ai X, Fine A. Ghosh S, et al. J Biol Chem. 2011 Jun 24;286(25):22678-87. doi: 10.1074/jbc.M111.241224. Epub 2011 May 2. J Biol Chem. 2011. PMID: 21536678 Free PMC article. - Notch Signaling in Vascular Smooth Muscle Cells.
Baeten JT, Lilly B. Baeten JT, et al. Adv Pharmacol. 2017;78:351-382. doi: 10.1016/bs.apha.2016.07.002. Epub 2016 Aug 26. Adv Pharmacol. 2017. PMID: 28212801 Free PMC article. Review. - The pericyte: A critical cell in the pathogenesis of CADASIL.
Ruchoux MM, Kalaria RN, Román GC. Ruchoux MM, et al. Cereb Circ Cogn Behav. 2021;2:100031. doi: 10.1016/j.cccb.2021.100031. Cereb Circ Cogn Behav. 2021. PMID: 34950895 Free PMC article. Review.
References
- Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ Res. 2005;97:512–523. - PubMed
- Jain RK. Molecular regulation of vessel maturation. Nat Med. 2003;9:685–693. - PubMed
- Montesano R, Pepper MS, Orci L. Paracrine induction of angiogenesis in vitro by Swiss 3T3 fibroblasts. J Cell Sci. 1993;105:1013–1024. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous