The subcellular organization of neocortical excitatory connections - PubMed (original) (raw)
The subcellular organization of neocortical excitatory connections
Leopoldo Petreanu et al. Nature. 2009.
Abstract
Understanding cortical circuits will require mapping the connections between specific populations of neurons, as well as determining the dendritic locations where the synapses occur. The dendrites of individual cortical neurons overlap with numerous types of local and long-range excitatory axons, but axodendritic overlap is not always a good predictor of actual connection strength. Here we developed an efficient channelrhodopsin-2 (ChR2)-assisted method to map the spatial distribution of synaptic inputs, defined by presynaptic ChR2 expression, within the dendritic arborizations of recorded neurons. We expressed ChR2 in two thalamic nuclei, the whisker motor cortex and local excitatory neurons and mapped their synapses with pyramidal neurons in layers 3, 5A and 5B (L3, L5A and L5B) in the mouse barrel cortex. Within the dendritic arborizations of L3 cells, individual inputs impinged onto distinct single domains. These domains were arrayed in an orderly, monotonic pattern along the apical axis: axons from more central origins targeted progressively higher regions of the apical dendrites. In L5 arborizations, different inputs targeted separate basal and apical domains. Input to L3 and L5 dendrites in L1 was related to whisker movement and position, suggesting that these signals have a role in controlling the gain of their target neurons. Our experiments reveal high specificity in the subcellular organization of excitatory circuits.
Figures
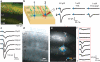
Figure 1. Subcellular Channelrhodopsin-2-Assisted Circuit Mapping (sCRACM)
a, Confocal image showing L2/3 neurons expressing mCherry (red) and ChR2-Venus (green) in the barrel cortex. b, Left, Schematic of the photostimulation geometry. Right, EPSCs evoked by photostimuli corresponding to the locations indicated in the schematic. Blue ticks indicate the laser pulse. Laser power is indicated on top. c, EPSCsCRACM evoked by photostimulation with increasing laser powers (right). d, Brightfield image of a brain slice showing the recording pipette and the photostimulation grid (blue dots). e, sCRACM map overlayed on a fluorescence image, showing ChR2-positive neurons, and the reconstructed dendrite of the recorded neuron (same experiment as d). Non-zero EPSCsCRACM are color-coded to represent mean amplitude. f, EPSCsCRACM recorded in the boxed regions in Panel e.
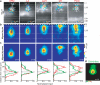
Figure 2. Subcellular distribution of inputs onto L3 pyramidal neurons
a, Examples of sCRACM maps overlaid on reconstructed dendrites and fluorescence images showing ChR2-positive axons (VPM, M1 and POm) or axons and dendrites (L2/3 and L4). White arrow head, bundle of ascending axons from M1. b, Group averages aligned by pia position (White triangles, soma position). c, top: Group averages aligned by soma position ; bottom: vertical profiles of the distribution of synaptic input (red) and the dendritic length density (green; from d). d, Average normalized dendritic length density of L3 pyramidal neurons. Error bars, s.e.m.
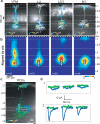
Figure 3. Subcellular distribution of inputs onto L5B pyramidal neurons
a, Examples of sCRACM maps overlaid on reconstructed dendritic arbors and fluorescence images. b, Group averages aligned by pia position. White triangles, soma position. c, sCRACM map of POm input onto a L5A pyramidal cell (blue). No responses were detected on the L5B neuron (green). d, EPSCsCRACM recorded on the L5A neuron (blue) or the L5B neuron (green) when photostimuating the boxed regions in Panel c . The stimulus occurred at the beginning of each trace.
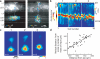
Figure 4. The laminar position of L5 pyramidal neurons determines the dendritic location of L2/3 inputs
a, Examples of the subcellular distribution of L2/3 input on superficial (L5A) (left) or deep (L5B) (right) pyramidal neurons. b, Vertical profiles of the subcellular distribution of L2/3→L5 inputs. Each column represents one cell, ordered by cortical depth. Cells were aligned by pia position. The relative density of L2/3 axons in the deep layers is indicated to the left of the panel. c, Average subcellular location of L2/3 input (aligned by soma position) of L5 cells grouped by increasing distance from the pia (groups correspond to the white lines in b).d, Plot of the vertical distance from the soma to the center of mass of L2/3 input on the perisomatic region ( < 285 μm from the soma) of L5 pyramidal neurons vs. cortical depth. The line is a regression fit.
Similar articles
- Parvalbumin-producing striatal interneurons receive excitatory inputs onto proximal dendrites from the motor thalamus in male mice.
Nakano Y, Karube F, Hirai Y, Kobayashi K, Hioki H, Okamoto S, Kameda H, Fujiyama F. Nakano Y, et al. J Neurosci Res. 2018 Jul;96(7):1186-1207. doi: 10.1002/jnr.24214. Epub 2018 Jan 4. J Neurosci Res. 2018. PMID: 29314192 - Organization of cortical and thalamic input to pyramidal neurons in mouse motor cortex.
Hooks BM, Mao T, Gutnisky DA, Yamawaki N, Svoboda K, Shepherd GM. Hooks BM, et al. J Neurosci. 2013 Jan 9;33(2):748-60. doi: 10.1523/JNEUROSCI.4338-12.2013. J Neurosci. 2013. PMID: 23303952 Free PMC article. - Local connections of excitatory neurons in motor-associated cortical areas of the rat.
Kaneko T. Kaneko T. Front Neural Circuits. 2013 May 28;7:75. doi: 10.3389/fncir.2013.00075. eCollection 2013. Front Neural Circuits. 2013. PMID: 23754982 Free PMC article. Review. - Inter- and intra-laminar connections of pyramidal cells in the neocortex.
Bannister AP. Bannister AP. Neurosci Res. 2005 Oct;53(2):95-103. doi: 10.1016/j.neures.2005.06.019. Neurosci Res. 2005. PMID: 16054257 Review.
Cited by
- Cell-type-specific integration of feedforward and feedback synaptic inputs in the posterior parietal cortex.
Rindner DJ, Proddutur A, Lur G. Rindner DJ, et al. Neuron. 2022 Nov 16;110(22):3760-3773.e5. doi: 10.1016/j.neuron.2022.08.019. Epub 2022 Sep 9. Neuron. 2022. PMID: 36087582 Free PMC article. - Target-specific M1 inputs to infragranular S1 pyramidal neurons.
Kinnischtzke AK, Fanselow EE, Simons DJ. Kinnischtzke AK, et al. J Neurophysiol. 2016 Sep 1;116(3):1261-74. doi: 10.1152/jn.01032.2015. Epub 2016 Jun 22. J Neurophysiol. 2016. PMID: 27334960 Free PMC article. - Callosal projections drive neuronal-specific responses in the mouse auditory cortex.
Rock C, Apicella AJ. Rock C, et al. J Neurosci. 2015 Apr 29;35(17):6703-13. doi: 10.1523/JNEUROSCI.5049-14.2015. J Neurosci. 2015. PMID: 25926449 Free PMC article. - Optogenetic Activation of Afferent Pathways in Brain Slices and Modulation of Responses by Volatile Anesthetics.
Murphy CA, Raz A, Grady SM, Banks MI. Murphy CA, et al. J Vis Exp. 2020 Jul 23;(161):10.3791/61333. doi: 10.3791/61333. J Vis Exp. 2020. PMID: 32773759 Free PMC article. - PyRhO: A Multiscale Optogenetics Simulation Platform.
Evans BD, Jarvis S, Schultz SR, Nikolic K. Evans BD, et al. Front Neuroinform. 2016 Mar 11;10:8. doi: 10.3389/fninf.2016.00008. eCollection 2016. Front Neuroinform. 2016. PMID: 27148037 Free PMC article.
References
- Shepherd GM, Pologruto TA, Svoboda K. Circuit analysis of experience-dependent plasticity in the developing rat barrel cortex. Neuron. 2003;38:277–89. - PubMed
- Williams SR, Stuart GJ. Site independence of EPSP time course is mediated by dendritic I(h) in neocortical pyramidal neurons. J Neurophysiol. 2000;83:3177–82. - PubMed
- Nevian T, Larkum ME, Polsky A, Schiller J. Properties of basal dendrites of layer 5 pyramidal neurons: a direct patch-clamp recording study. Nat Neurosci. 2007;10:206–14. - PubMed
- Stepanyants A, Hof PR, Chklovskii DB. Geometry and structural plasticity of synaptic connectivity. Neuron. 2002;34:275–88. - PubMed
- Douglas RJ, Martin KA. Mapping the matrix: the ways of neocortex. Neuron. 2007;56:226–38. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials