Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells - PubMed (original) (raw)
Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells
Bo Wen et al. Nat Genet. 2009 Feb.
Abstract
Higher eukaryotes must adapt a totipotent genome to specialized cell types with stable but limited functions. One potential mechanism for lineage restriction is changes in chromatin, and differentiation-related chromatin changes have been observed for individual genes. We have taken a genome-wide view of histone H3 lysine 9 dimethylation (H3K9Me2) and find that differentiated tissues show surprisingly large K9-modified regions (up to 4.9 Mb). These regions are highly conserved between human and mouse and are differentiation specific, covering only approximately 4% of the genome in undifferentiated mouse embryonic stem (ES) cells, compared to 31% in differentiated ES cells, approximately 46% in liver and approximately 10% in brain. These modifications require histone methyltransferase G9a and are inversely related to expression of genes within the regions. We term these regions large organized chromatin K9 modifications (LOCKs). LOCKs are substantially lost in cancer cell lines, and they may provide a cell type-heritable mechanism for phenotypic plasticity in development and disease.
Figures
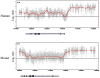
Figure 1. LOCK-like clustering of histone H3 lysine-9 dimethylation (H3K9Me2) in human placenta
Freshly purified native chromatin digested with micrococcal nuclease was immunoprecipitated with antibody to H3K9Me2 and hybridized to human genomic tiling arrays (ChIP-on-chip), revealing two LOCK-like clusters of ~2 Mb each in the chromosome 15 Prader-Willi/Angelman syndromes imprinted gene region. The X axis represents genomic positions and the Y axis gives log2 ratios of ChIP over input signal. The gray dots represent individual probe data, and the red line denotes the smoothed curve by sliding windows. The orange bars in the middle show locations of known CTCF binding sites and gene annotations are given on the bottom. The paternally expressed genes (Mkrn3, Ndn, Snrpn and Gabrb3) are localized within and near the edges of LOCK, whereas the maternally expressed genes (Ube3a and Atp10a) are localized between the LOCKs. CTCF binding sites are largely located at the boundaries of the LOCKs.
Figure 2. H3K9Me2 LOCKs are conserved between human and mouse
ChIP-on-chip data of the ENCODE region ENr213 in human placenta and its syntenic region in mouse liver. Dots and curves are as in Fig. 1. The location of H3K9Me2 LOCKs is highly consistent between human and mouse. The LOCK on the right part of the figure begins at the 3′end of the CDH2 gene in both human and mouse. See also Supplementary Fig. 4.
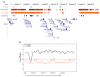
Figure 3. H3K9Me2 LOCKs arise during differentiation
a, Depicted is a summary of data from a 10 Mb region of mouse chromosome 8. Locations of LOCKs in undifferentiated mouse embryonic stem (ES) cells, differentiated ES cells, liver and brain are denoted by green, red, orange and blue bars, respectively. b, LOCKs are almost completely absent in differentiated G9a knockout ES cells, compared to wild type ES cells. The black and red lines represent smoothed curves of H3K9Me2 in differentiated WT and G9a knockout ES cells, respectively. See also Supplementary Fig 6.
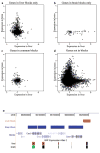
Figure 4. Tissue-specific H3K9Me2 LOCKs are associated with differential gene expression in liver and brain
Levels of gene expression in liver (X axis) and brain (Y axis) of genes in liver LOCKs only (a), genes in brain LOCKs only (b), genes in LOCKs common to liver and brain (c) and genes not in LOCKs in liver or brain (d). Genes that lay within LOCKs in the liver but not in the brain were largely silenced in liver, but showed a broad range of expression in brain (P = 0.002). Similarly, genes in LOCKs in brain but not liver, while fewer in number were largely silenced in brain, but showed a broad range of expression in liver (P = 10−9). Genes that lay within LOCKs in both tissues were silenced in both, while genes not in LOCKs in either liver or brain were broadly expressed in both tissues (P = 0.0002 and 0.005 for liver and brain, respectively). e, an example of a gene clusters in a tissue-specific LOCK. Expression levels of transcripts are based on the GNF Atlas track from UCSC genome browser. This region is marked by brain LOCKs, and correspondingly, the genes under LOCKs are silenced (green) in brain but highly expressed (red) in liver.
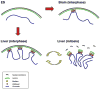
Figure 5. A model of epigenetic memory of cell type-specific higher-order chromatin structure mediated by H3K9Me2 LOCKs
In undifferentiated ES cells (upper left), the genome is largely unmarked by H3K9Me2 LOCKs, chromosomal positioning is relatively apart from the nuclear membrane and shows global expression. In differentiated cells such as brain (upper right) and liver (lower left), large fractions of the genome are marked by H3K9Me2 LOCKs in a tissue-specific manner. The chromosome may be repositioned through the association of H3K9Me2 and nuclear lamina through unknown mediator proteins, forming lineage-specific nuclear architectures. During cell division, the lamina-LOCK association can be remembered in mitosis (lower right) and the nuclear architecture reestablished through a mechanism dependent on nuclear envelope breakdown and reformation, and enhanced by contiguous sites of H3K9Me2 modification.
Comment in
- Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells.
Filion GJ, van Steensel B. Filion GJ, et al. Nat Genet. 2010 Jan;42(1):4; author reply 5-6. doi: 10.1038/ng0110-4. Nat Genet. 2010. PMID: 20037608 No abstract available.
Similar articles
- G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis.
Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. Tachibana M, et al. Genes Dev. 2002 Jul 15;16(14):1779-91. doi: 10.1101/gad.989402. Genes Dev. 2002. PMID: 12130538 Free PMC article. - Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9.
Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Tachibana M, et al. Genes Dev. 2005 Apr 1;19(7):815-26. doi: 10.1101/gad.1284005. Epub 2005 Mar 17. Genes Dev. 2005. PMID: 15774718 Free PMC article. - Histone H3-K9 methyltransferase ESET is essential for early development.
Dodge JE, Kang YK, Beppu H, Lei H, Li E. Dodge JE, et al. Mol Cell Biol. 2004 Mar;24(6):2478-86. doi: 10.1128/MCB.24.6.2478-2486.2004. Mol Cell Biol. 2004. PMID: 14993285 Free PMC article. - Dynamic protein methylation in chromatin biology.
Ng SS, Yue WW, Oppermann U, Klose RJ. Ng SS, et al. Cell Mol Life Sci. 2009 Feb;66(3):407-22. doi: 10.1007/s00018-008-8303-z. Cell Mol Life Sci. 2009. PMID: 18923809 Free PMC article. Review. - Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human.
Malik S, Bhaumik SR. Malik S, et al. FEBS J. 2010 Apr;277(8):1805-21. doi: 10.1111/j.1742-4658.2010.07607.x. Epub 2010 Mar 4. FEBS J. 2010. PMID: 20236312 Free PMC article. Review.
Cited by
- Dynamic distribution of linker histone H1.5 in cellular differentiation.
Li JY, Patterson M, Mikkola HK, Lowry WE, Kurdistani SK. Li JY, et al. PLoS Genet. 2012;8(8):e1002879. doi: 10.1371/journal.pgen.1002879. Epub 2012 Aug 30. PLoS Genet. 2012. PMID: 22956909 Free PMC article. - Influences of lamin A levels on induction of pluripotent stem cells.
Zuo B, Yang J, Wang F, Wang L, Yin Y, Dan J, Liu N, Liu L. Zuo B, et al. Biol Open. 2012 Nov 15;1(11):1118-27. doi: 10.1242/bio.20121586. Epub 2012 Sep 7. Biol Open. 2012. PMID: 23213392 Free PMC article. - Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host.
Timp W, Feinberg AP. Timp W, et al. Nat Rev Cancer. 2013 Jul;13(7):497-510. doi: 10.1038/nrc3486. Epub 2013 Jun 13. Nat Rev Cancer. 2013. PMID: 23760024 Free PMC article. Review. - Joint annotation of chromatin state and chromatin conformation reveals relationships among domain types and identifies domains of cell-type-specific expression.
Libbrecht MW, Ay F, Hoffman MM, Gilbert DM, Bilmes JA, Noble WS. Libbrecht MW, et al. Genome Res. 2015 Apr;25(4):544-57. doi: 10.1101/gr.184341.114. Epub 2015 Feb 12. Genome Res. 2015. PMID: 25677182 Free PMC article. - Programmed suppression of oxidative phosphorylation and mitochondrial function by gestational alcohol exposure correlate with widespread increases in H3K9me2 that do not suppress transcription.
Chang RC, Thomas KN, Mehta NA, Veazey KJ, Parnell SE, Golding MC. Chang RC, et al. Epigenetics Chromatin. 2021 Jun 15;14(1):27. doi: 10.1186/s13072-021-00403-w. Epigenetics Chromatin. 2021. PMID: 34130715 Free PMC article.
References
- Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–71. - PubMed
- Atkinson SP, et al. Epigenetic marking prepares the human HOXA cluster for activation during differentiation of pluripotent cells. Stem Cells. 2008 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 RR021967/RR/NCRR NIH HHS/United States
- R37 CA054358/CA/NCI NIH HHS/United States
- P50 HG003233/HG/NHGRI NIH HHS/United States
- P50 HG003233-05/HG/NHGRI NIH HHS/United States
- R01 GM083084/GM/NIGMS NIH HHS/United States
- R37 CA054358-18/CA/NCI NIH HHS/United States
- R01 CA054358/CA/NCI NIH HHS/United States
- R01 GM083084-02/GM/NIGMS NIH HHS/United States
- R37 CA054358-19/CA/NCI NIH HHS/United States
- R01 RR021967-02/RR/NCRR NIH HHS/United States
- P50HG003233/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous