ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1-mediated pathway - PubMed (original) (raw)
ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1-mediated pathway
Bo Hu et al. Cancer Res. 2009.
Abstract
A common pathobiological feature of malignant gliomas is the insidious infiltration of single tumor cells into the brain parenchyma, rendering these deadly tumors virtually incurable with available therapies. In this study, we report that ADP-ribosylation factor 6 (ARF6), a Ras superfamily small GTPase, is abundantly expressed in invasive human glioma cells. Cellular depletion of ARF6 by small interfering RNA decreased Rac1 activation, impaired HGF-stimulated and serum-stimulated glioma cell migration in vitro, and markedly decreased the invasive capacity of invasive glioma in the brain. Furthermore, ectopic expression of ARF6 in glioma cells promoted cell migration via the activation of Rac1. Upon stimulation of glioma cells with HGF, we show that IQ-domain GTPase-activating protein 1 (IQGAP1) is recruited and overlaps with ARF6 at the leading edge of migrating cells. However, cellular depletion of ARF6 abrogated this recruitment of IQGAP1 and attenuated the formation of surface protrusions. ARF6 forms complexes with Rac1 and IQGAP1 in glioma cells upon HGF stimulation, and knockdown of IQGAP1 significantly inhibits ARF6-induced Rac1 activation and cell migration. Taken together, these data suggest that ARF6-mediated Rac1 activation is essential for glioma cell invasion via a signaling pathway that requires IQGAP1.
Figures
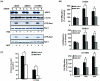
Figure 1. Suppression of ARF6 by siRNA inhibits glioma cell migration in vitro and cell invasion ex vivo
(A), IB analyses of siRNA knockdown of ARF6 and the impact on Rac1 activation by FBS and HGF in indicated glioma cells. LN229, SNB19, and U373MG cells were transiently transfected with ARF6 siRNA or a control siRNA for 48 hr in media containing 10% FBS. Afterwards, the transfected cells were cultured in media containing no serum, 10% FBS or 20 ng/ml HGF for an additional 24 hr followed by IB with an anti-ARF6 antibody and analyses of GTP loading of Rac1 using a Rac1 activation assay kit. IB for β-actin and Rac1 were used as protein loading controls. Data are representative of three independent experiments with similar results. (B), In vitro migration assays. LN229, SNB19, and U373MG cells were transiently transfected with ARF6 siRNA or control siRNA, followed by in vitro cell migration assays. Fifty μl of various siRNA-transfected cells in serum-free (SF) DMEM media containing 0.05% BSA was separately placed into the top compartment of a Boyden Chamber. Serum-free DMEM media containing 0.05% BSA with or without 20 ng/ml HGF or 10% FBS were placed in the bottom wells. The cells were allowed to migrate through an 8 μm-pore size membrane pre-coated with gelatin (10 μg/ml) for 16 hr at 37°C. Cell migration was determined as previously described (16). Control, media without serum or HGF. Columns, mean percentage of migrating control cells from three independent experiments in six replicates per pair per cell line (control siRNA-transfected and ARF6-siRNA-transfected cells); bars, SD. *, p < 0.05, one-way ANOVA followed by Newman-Keuls post hoc. (C), GFP-expressing SNB19 and U373MG cells were transiently transfected with ARF6 siRNA or control siRNA, followed by an ex vivo brain slice invasion assay. Depth of the GFP-expressing glioma cell invasion into a murine brain slice was determined by optical sectioning using a Zeiss LSM 510 confocal microscope. Columns, mean distance (μm) invaded in 48 hr from six independent experiments in six replicates per pair (control siRNA-transfected and ARF6-siRNA-transfected cells); bars, SD. *, p < 0.05, one-way ANOVA followed by Newman-Keuls post hoc.
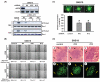
Figure 2. Suppression of ARF6 by siRNA inhibits glioma cell migration in vitro, and invasion ex vivo and in vivo
(A), SNB19 cells were stably transfected with ARF6 siRNA or control siRNA and cultured in media containing no serum, 10% FBS or 20 ng/ml HGF for 24 hr. Inhibition of ARF6 proteins in the G418-resistent cell clones (control, ARF6-siRNA #4, #10, and #12) and the impact on GTP loading of Rac1 were analyzed by IB with an anti-ARF6 antibody and a Rac1 activation assay kit. IB for β-actin and Rac1 were used as protein loading controls. (B), Cell motility of ARF6-siRNA or control-siRNA cell clones in the presence of 10% FBS or 20 ng/ml HGF was evaluated by an in vitro wound healing assay. Quantification of cell migration (averaging the position of the migrating cells at the wounding edges) in the monolayer wound healing assays are shown under the images as the means values (± SE in μm) of eight measurements at a 24-hr time point for each condition. Bar = 100 μm. (C), Cell invasion of ARF6-siRNA or control-siRNA cell clones was determined by an ex vivo brain slice invasion assay. Top panels, representative epifluorescent images of the GFP-expressing SNB19 ARF6-siRNA or control-siRNA cell clones were captured using a digital camera attached to a stereomicroscope at 40X magnification. White arrows indicate lateral cell migration and invasion 48 hr after initial placement of indicated cells onto the top of brain slices. Bar graph shows invasion depth of ARF6-siRNA or control-siRNA cell clones into a murine brain slice measured by confocal microscopy. Columns, mean distance (μm) invaded in 48 hr from four independent experiments done in six replicates per pair (ARF6-siRNA vs control-siRNA cell clones). Bars, SE. *, P < 0.05, one-way ANOVA followed by Newman-Keuls post hoc. No transfection controls (no siRNA) for SNB19 cells were also done showing no observable effects on cell viability or the invasive ability when comparing the control siRNA and non-transfected cells (data not shown). Data shown in (A) to (C) were from three independent experiments with triplicate (in vitro) and six brain tissue slices (ex vivo) with similar results. (D) Intracranial gliomas established by SNB19 control-siRNA cells (panels a and d), ARF6-siRNA #10 cells (b and e), and ARF6-siRNA #12 cells (c and f) in the murine brains were analyzed by H&E staining (a to c) and epifluorescent images (e to f). Arrows, invasive extensions as well as disseminated tumor clusters. Arrowheads, the clean edge of tumors. The animal experiments were done two independent times with six mice per group with similar results.
Figure 3. ARF6 is not required for HGF-stimulated protein phosphorylation of ERK1/2 in glioma cells
Human glioma LN229, SNB19 and U373MG cell lines were transfected with siRNA for ARF6 and control then treated with 20 ng/ml of HGF for 20 min. Inhibition of ARF6 proteins in transfected cells and the impacts on ERK1/2 protein phosphorylation and GTP loading of Rac1 were analyzed with IB using anti-phospho-ERK1/2, anti-ERK1/2 and anti-ARF6 antibodies, and a Rac1 activation assay kit. IB for β-actin and Rac1 were used as protein loading controls. Data are representative of three independent experiments with similar results.
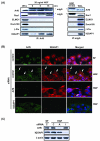
Figure 4. ARF6 is associated with IQGAP1 in glioma cells and controls formation of extended ruffles on plasma membrane
(A), ARF6 is associated with IQGAP1 in glioma cells. The serum-starved SNB19 cells were treated with 20 ng/ml of HGF for indicated times and lysed. The cell lysates were immunoprecipitated (IP) with an anti-ARF6 antibody (left panels) or an isotype-matched mouse IgG control (right panels) followed by IB separately using anti-ARF6, anti-Rac1, anti-Dock180, anti-ELMO1, anti-β-PIX and anti-IQGAP1 antibodies. IB for whole cell lysates (WCL) was used as a protein input control. (B), SNB19 cells were transiently transfected with ARF6 siRNA, IQGAP1 siRNA or a control siRNA, and then under serum starvation for 48 hr followed by HGF treatment (20 ng/ml) for 30 min. Co-localization of ARF6 and IQGAP1 at the invasion fronts of cells was determined by immunofluorescent staining of various cells using anti-ARF6 and anti-IQGAP1 antibodies. Nuclei were stained by Hoechst (blue). Images were captured using an Olympus BX51 microscope equipped with a digital camera (magnification: 400X). Arrows in the second panels indicate co-localization of ARF6 and IQGAP1 at the invasion fronts. Percentage of cells showing the co-localization was estimated by a blind observer (J.-J. Yiin) in 10 random fields at 10X magnification and is described in the text. (C), IB analyses of siRNA knockdown of ARF6 and IQGAP1 in SNB19 cells. Whole cell lysates of a portion of SNB19 cells that were immunostained in (B) were analyzed by IB for inhibition of ARF6 and IQGAP1 expression by ARF6 siRNA, IQGAP1 siRNA or a control siRNA using anti-ARF6 and anti-IQGAP1 antibodies. IB for β-actin was used as a protein loading control. Data shown in (A) to (C) were from three independent experiments with similar results.
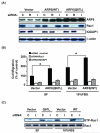
Figure 5. IQGAP1 is necessary for the signal connection between ARF6 and Rac1, and ARF6-mediated glioma cell migration
(A), IB analyses of siRNA knockdown of Rac1 or IQGAP1 in various U87MG glioma cells. U87MG cells with low levels of endogenous ARF6 were transiently transfected with ARF6-WT, ARF6-Q67L, or vector only. G418-resistant U87MG, U87MG/ARF6-WT, and U87MG/ARF6-Q67L cell populations were then transiently transfected with control siRNA, Rac1 siRNA, and IQGAP1 siRNA followed by IB with anti-ARF6, anti-Rac1, and anti-IQGAP1 antibodies. IB for β-actin was used as a protein loading control. (B), Impact of knocking down IQGAP1 or Rac1 on cell motility of various indicated cells was determined by in vitro migration assays under conditions of serum-free (SF) and 10% FBS. (C), Effect of inhibition of IQGAP1 on activation of Rac1 in various indicated cells in the absence or presence of 10% FBS was examined using a Rac1 activation assay kit under conditions of serum-free (SF) and 10% FBS. Whole cell lysates were prepared from a portion of various IQGAP1 siRNA-transfected U87MG cells shown in (A) that were cultured in media with or without 10% FBS for an additional 24 hrs after siRNA transient transfection. Expression of endogenous IQGAP1 proteins and the inhibition of IQGAP1 expression by siRNA knockdown in various U87MG cells cultured in media containing 10% FBS are shown in the third panel of IB analysis in (A). IB for Rac1 was used as protein loading controls. Data shown in (A) to (C) are representative from three independent experiments with similar results.
Similar articles
- GEP100/Arf6 is required for epidermal growth factor-induced ERK/Rac1 signaling and cell migration in human hepatoma HepG2 cells.
Hu Z, Du J, Yang L, Zhu Y, Yang Y, Zheng D, Someya A, Gu L, Lu X. Hu Z, et al. PLoS One. 2012;7(6):e38777. doi: 10.1371/journal.pone.0038777. Epub 2012 Jun 11. PLoS One. 2012. PMID: 22701712 Free PMC article. - ARF1 and ARF6 regulate recycling of GRASP/Tamalin and the Rac1-GEF Dock180 during HGF-induced Rac1 activation.
Koubek EJ, Santy LC. Koubek EJ, et al. Small GTPases. 2018 May 4;9(3):242-259. doi: 10.1080/21541248.2016.1219186. Epub 2016 Aug 26. Small GTPases. 2018. PMID: 27562622 Free PMC article. - ARF6 promotes the formation of Rac1 and WAVE-dependent ventral F-actin rosettes in breast cancer cells in response to epidermal growth factor.
Marchesin V, Montagnac G, Chavrier P. Marchesin V, et al. PLoS One. 2015 Mar 23;10(3):e0121747. doi: 10.1371/journal.pone.0121747. eCollection 2015. PLoS One. 2015. PMID: 25799492 Free PMC article. - Roles of Arf6 in cancer cell invasion, metastasis and proliferation.
Li R, Peng C, Zhang X, Wu Y, Pan S, Xiao Y. Li R, et al. Life Sci. 2017 Aug 1;182:80-84. doi: 10.1016/j.lfs.2017.06.008. Epub 2017 Jun 15. Life Sci. 2017. PMID: 28625359 Review. - New model for the interaction of IQGAP1 with CDC42 and RAC1.
Nouri K, Timson DJ, Ahmadian MR. Nouri K, et al. Small GTPases. 2020 Jan;11(1):16-22. doi: 10.1080/21541248.2017.1321169. Epub 2017 Jun 19. Small GTPases. 2020. PMID: 28622072 Free PMC article. Review.
Cited by
- A role for activated Cdc42 in glioblastoma multiforme invasion.
Okura H, Golbourn BJ, Shahzad U, Agnihotri S, Sabha N, Krieger JR, Figueiredo CA, Chalil A, Landon-Brace N, Riemenschneider A, Arai H, Smith CA, Xu S, Kaluz S, Marcus AI, Van Meir EG, Rutka JT. Okura H, et al. Oncotarget. 2016 Aug 30;7(35):56958-56975. doi: 10.18632/oncotarget.10925. Oncotarget. 2016. PMID: 27486972 Free PMC article. - Tumour-intrinsic endomembrane trafficking by ARF6 shapes an immunosuppressive microenvironment that drives melanomagenesis and response to checkpoint blockade therapy.
Wee Y, Wang J, Wilson EC, Rich CP, Rogers A, Tong Z, DeGroot E, Gopal YNV, Davies MA, Ekiz HA, Tay JKH, Stubben C, Boucher KM, Oviedo JM, Fairfax KC, Williams MA, Holmen SL, Wolff RK, Grossmann AH. Wee Y, et al. Nat Commun. 2024 Aug 4;15(1):6613. doi: 10.1038/s41467-024-50881-1. Nat Commun. 2024. PMID: 39098861 Free PMC article. - Nucleotide exchange factors: Kinetic analyses and the rationale for studying kinetics of GEFs.
Northup JK, Jian X, Randazzo PA. Northup JK, et al. Cell Logist. 2012 Jul 1;2(3):140-146. doi: 10.4161/cl.21627. Cell Logist. 2012. PMID: 23181196 Free PMC article. - The small GTPase ARF6 stimulates β-catenin transcriptional activity during WNT5A-mediated melanoma invasion and metastasis.
Grossmann AH, Yoo JH, Clancy J, Sorensen LK, Sedgwick A, Tong Z, Ostanin K, Rogers A, Grossmann KF, Tripp SR, Thomas KR, D'Souza-Schorey C, Odelberg SJ, Li DY. Grossmann AH, et al. Sci Signal. 2013 Mar 5;6(265):ra14. doi: 10.1126/scisignal.2003398. Sci Signal. 2013. PMID: 23462101 Free PMC article. - CD13 tethers the IQGAP1-ARF6-EFA6 complex to the plasma membrane to promote ARF6 activation, β1 integrin recycling, and cell migration.
Ghosh M, Lo R, Ivic I, Aguilera B, Qendro V, Devarakonda C, Shapiro LH. Ghosh M, et al. Sci Signal. 2019 Apr 30;12(579):eaav5938. doi: 10.1126/scisignal.aav5938. Sci Signal. 2019. PMID: 31040262 Free PMC article.
References
- Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. - PubMed
- Giese A, Bjerkvig R, Berens ME, et al. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21:1624–36. - PubMed
- D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–58. - PubMed
- Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem. 2003;278:41573–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA102011/CA/NCI NIH HHS/United States
- R01 CA130966/CA/NCI NIH HHS/United States
- R01 CA115316/CA/NCI NIH HHS/United States
- CA102011/CA/NCI NIH HHS/United States
- CA115316/CA/NCI NIH HHS/United States
- R56 CA115316/CA/NCI NIH HHS/United States
- CA130966/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous