Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data - PubMed (original) (raw)
Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data
Anton Valouev et al. Nat Methods. 2008 Sep.
Abstract
Molecular interactions between protein complexes and DNA mediate essential gene-regulatory functions. Uncovering such interactions by chromatin immunoprecipitation coupled with massively parallel sequencing (ChIP-Seq) has recently become the focus of intense interest. We here introduce quantitative enrichment of sequence tags (QuEST), a powerful statistical framework based on the kernel density estimation approach, which uses ChIP-Seq data to determine positions where protein complexes contact DNA. Using QuEST, we discovered several thousand binding sites for the human transcription factors SRF, GABP and NRSF at an average resolution of about 20 base pairs. MEME motif-discovery tool-based analyses of the QuEST-identified sequences revealed DNA binding by cofactors of SRF, providing evidence that cofactor binding specificity can be obtained from ChIP-Seq data. By combining QuEST analyses with Gene Ontology (GO) annotations and expression data, we illustrate how general functions of transcription factors can be inferred.
Figures
Figure 1
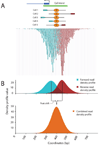
QuEST’s representation of ChIP-Seq data using density profiles.. (A) GABP ChIP-Seq reads from the promoter and CpG island of the Nitric oxide synthase interacting protein gene. Hypothetical GABP binding in five cells and the corresponding DNA fragments with sequencing reads. Below, actual read data. Forward reads are displayed as small blue bands and reverse reads as small maroon bands. (B) Forward (blue) and reverse (maroon) Read Density Profiles derived from the read data contribute to the Combined Density Profile (orange). The zero x-coordinate corresponds to coordinate 54775300 of human Chromosome 19, NCBI build 36.
Figure 2
Reproducibility and robustness of QuEST results assessed by comparison between two independent NRSF data sets. (A) Correlation between NRSF polyclonal and NRSF monoclonal peak scores (rho = 0.97) with the inset expanding the portion near the graph origin. (B) Bar chart of the distance between NRSF polyclonal and NRSF monoclonal peak call positions.
Figure 3
Resolution of QuEST as quantified by the distance between QuEST peak calls and TFBS motif centers. Histograms in each panel represent the distribution of peak distances to the nearest high-scoring motif.
Figure 4
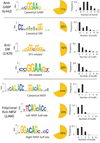
Motif analysis results. Each panel displays significantly overrepresented motif Weblogos for each of the three transcription factors. Pie-charts show the fraction of peaks with motifs in close proximity to the peak (< 100 bps). Histograms show the distribution of the motif number within 100 bps of the peak.
Similar articles
- Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data.
Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Jothi R, et al. Nucleic Acids Res. 2008 Sep;36(16):5221-31. doi: 10.1093/nar/gkn488. Epub 2008 Aug 6. Nucleic Acids Res. 2008. PMID: 18684996 Free PMC article. - De novo motif identification improves the accuracy of predicting transcription factor binding sites in ChIP-Seq data analysis.
Boeva V, Surdez D, Guillon N, Tirode F, Fejes AP, Delattre O, Barillot E. Boeva V, et al. Nucleic Acids Res. 2010 Jun;38(11):e126. doi: 10.1093/nar/gkq217. Epub 2010 Apr 7. Nucleic Acids Res. 2010. PMID: 20375099 Free PMC article. - MEME-ChIP: motif analysis of large DNA datasets.
Machanick P, Bailey TL. Machanick P, et al. Bioinformatics. 2011 Jun 15;27(12):1696-7. doi: 10.1093/bioinformatics/btr189. Epub 2011 Apr 12. Bioinformatics. 2011. PMID: 21486936 Free PMC article. - Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond.
Mundade R, Ozer HG, Wei H, Prabhu L, Lu T. Mundade R, et al. Cell Cycle. 2014;13(18):2847-52. doi: 10.4161/15384101.2014.949201. Cell Cycle. 2014. PMID: 25486472 Free PMC article. Review. - Genome Wide Approaches to Identify Protein-DNA Interactions.
Ma T, Ye Z, Wang L. Ma T, et al. Curr Med Chem. 2019;26(42):7641-7654. doi: 10.2174/0929867325666180530115711. Curr Med Chem. 2019. PMID: 29848263 Review.
Cited by
- Uncovering transcription factor modules using one- and three-dimensional analyses.
Lan X, Farnham PJ, Jin VX. Lan X, et al. J Biol Chem. 2012 Sep 7;287(37):30914-21. doi: 10.1074/jbc.R111.309229. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952238 Free PMC article. Review. - ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions.
Furey TS. Furey TS. Nat Rev Genet. 2012 Dec;13(12):840-52. doi: 10.1038/nrg3306. Epub 2012 Oct 23. Nat Rev Genet. 2012. PMID: 23090257 Free PMC article. Review. - On the immortality of television sets: "function" in the human genome according to the evolution-free gospel of ENCODE.
Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, Elhaik E. Graur D, et al. Genome Biol Evol. 2013;5(3):578-90. doi: 10.1093/gbe/evt028. Genome Biol Evol. 2013. PMID: 23431001 Free PMC article. - Genome-wide identification and annotation of HIF-1α binding sites in two cell lines using massively parallel sequencing.
Tanimoto K, Tsuchihara K, Kanai A, Arauchi T, Esumi H, Suzuki Y, Sugano S. Tanimoto K, et al. Hugo J. 2010 Dec;4(1-4):35-48. doi: 10.1007/s11568-011-9150-9. Epub 2011 Feb 19. Hugo J. 2010. PMID: 22132063 Free PMC article. - Next-generation sequencing for cancer diagnostics: a practical perspective.
Meldrum C, Doyle MA, Tothill RW. Meldrum C, et al. Clin Biochem Rev. 2011 Nov;32(4):177-95. Clin Biochem Rev. 2011. PMID: 22147957 Free PMC article.
References
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, Wheeler R, Wong B, Drenkow J, Yamanaka M, Patel S, Brubaker S, Tammana H, Helt G, Struhl K, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004 Feb 20;116(4):499–509. 2004. - PubMed
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated Signal Transduction Kinases Frequently Occupy Target Genes. Science. 2006 Jul 28;Vol. 313.(no. 5786):533–536. - PubMed
- Lieb JD. Genome-wide mapping of protein-DNA interactions by chromatin immunoprecipitation and DNA microarray hybridization. Methods Mol Biol. 2003;224:99–109. - PubMed
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007 Jun 8;316(5830):1497–1502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 HG003162/HG/NHGRI NIH HHS/United States
- U54 HG004576/HG/NHGRI NIH HHS/United States
- 5 U01 HG003162/HG/NHGRI NIH HHS/United States
- 1 U54-HG004576/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous