Biophysical characterization of intrinsically disordered proteins - PubMed (original) (raw)
Review
Biophysical characterization of intrinsically disordered proteins
David Eliezer. Curr Opin Struct Biol. 2009 Feb.
Abstract
The challenges associated with the structural characterization of disordered proteins have resulted in the application of a host of biophysical methods to such systems. NMR spectroscopy is perhaps the most readily suited technique for providing high-resolution structural information on disordered protein states in solution. Optical methods, solid state NMR, ESR and X-ray scattering can also provide valuable information regarding the ensemble of conformations sampled by disordered states. Finally, computational studies have begun to assume an increasingly important role in interpreting and extending the impact of experimental data obtained for such systems. This article discusses recent advances in the applications of these methods to intrinsically disordered proteins.
Conflict of interest statement
Conflict of Interest
The author has no conflicts to declare.
Figures
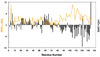
Figure 1. NMR α-carbon secondary chemical shifts and NH RDCs for β-synuclein
Chemical shift deviations from random coil values indicate a preference for extended structure (negative values) in the C-terminal region and helical structure (positive values) in the N-terminal region. Large amplitude RDCs in the C-terminal region correlate qualitatively with negative shift deviations and likely reflect locally extended chain segments. Smaller amplitude RDCs in the N-terminal region occur largely in regions of positive shift deviations, although the RDCs do not change sign as would be expected for more highly populated helical structure. Adapted from [26].
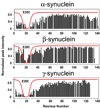
Figure 2. NMR PRE in α-, β- and γ-synuclein
A paramagnetic spin label attached to a cysteine introduced at position 20 leads to clear broadening in the C-terminal tail of α-synuclein, but not of β- or γ-synuclein, suggesting long-range N- to C-terminal contacts are present in the former but not in the latter. The red lines represent the predicted broadening for an idealized random coil. Adapted from [26].
Similar articles
- Atomic-level characterization of disordered protein ensembles.
Mittag T, Forman-Kay JD. Mittag T, et al. Curr Opin Struct Biol. 2007 Feb;17(1):3-14. doi: 10.1016/j.sbi.2007.01.009. Epub 2007 Jan 23. Curr Opin Struct Biol. 2007. PMID: 17250999 Review. - What can solid state NMR contribute to our understanding of protein folding?
Hu KN, Tycko R. Hu KN, et al. Biophys Chem. 2010 Sep;151(1-2):10-21. doi: 10.1016/j.bpc.2010.05.009. Epub 2010 May 23. Biophys Chem. 2010. PMID: 20542371 Free PMC article. Review. - Ensemble modeling of protein disordered states: experimental restraint contributions and validation.
Marsh JA, Forman-Kay JD. Marsh JA, et al. Proteins. 2012 Feb;80(2):556-72. doi: 10.1002/prot.23220. Epub 2011 Nov 17. Proteins. 2012. PMID: 22095648 - Describing intrinsically disordered proteins at atomic resolution by NMR.
Jensen MR, Ruigrok RW, Blackledge M. Jensen MR, et al. Curr Opin Struct Biol. 2013 Jun;23(3):426-35. doi: 10.1016/j.sbi.2013.02.007. Epub 2013 Mar 29. Curr Opin Struct Biol. 2013. PMID: 23545493 Review. - Insights into the conformations and dynamics of intrinsically disordered proteins using single-molecule fluorescence.
Gomes GN, Gradinaru CC. Gomes GN, et al. Biochim Biophys Acta Proteins Proteom. 2017 Nov;1865(11 Pt B):1696-1706. doi: 10.1016/j.bbapap.2017.06.008. Epub 2017 Jun 16. Biochim Biophys Acta Proteins Proteom. 2017. PMID: 28625737 Review.
Cited by
- A preformed binding interface in the unbound ensemble of an intrinsically disordered protein: evidence from molecular simulations.
Knott M, Best RB. Knott M, et al. PLoS Comput Biol. 2012;8(7):e1002605. doi: 10.1371/journal.pcbi.1002605. Epub 2012 Jul 19. PLoS Comput Biol. 2012. PMID: 22829760 Free PMC article. - bHLH-PAS Proteins: Their Structure and Intrinsic Disorder.
Kolonko M, Greb-Markiewicz B. Kolonko M, et al. Int J Mol Sci. 2019 Jul 26;20(15):3653. doi: 10.3390/ijms20153653. Int J Mol Sci. 2019. PMID: 31357385 Free PMC article. Review. - Detection of transient interchain interactions in the intrinsically disordered protein alpha-synuclein by NMR paramagnetic relaxation enhancement.
Wu KP, Baum J. Wu KP, et al. J Am Chem Soc. 2010 Apr 28;132(16):5546-7. doi: 10.1021/ja9105495. J Am Chem Soc. 2010. PMID: 20359221 Free PMC article. - E46K Parkinson's-linked mutation enhances C-terminal-to-N-terminal contacts in alpha-synuclein.
Rospigliosi CC, McClendon S, Schmid AW, Ramlall TF, Barré P, Lashuel HA, Eliezer D. Rospigliosi CC, et al. J Mol Biol. 2009 May 22;388(5):1022-32. doi: 10.1016/j.jmb.2009.03.065. Epub 2009 Apr 5. J Mol Biol. 2009. PMID: 19345692 Free PMC article. - Insights into the Function of the Unstructured N-Terminal Domain of Proteins 4.1R and 4.1G in Erythropoiesis.
Nunomura W, Gascard P, Takakuwa Y. Nunomura W, et al. Int J Cell Biol. 2011;2011:943272. doi: 10.1155/2011/943272. Epub 2011 Aug 28. Int J Cell Biol. 2011. PMID: 21904552 Free PMC article.
References
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. - PubMed
- Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. - PubMed
- Tompa P, Dosztanyi Z, Simon I. Prevalent structural disorder in E. coli and S. cerevisiae proteomes. J Proteome Res. 2006;5:1996–2000. - PubMed
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. - PubMed
- Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG019391/AG/NIA NIH HHS/United States
- R37 AG019391/AG/NIA NIH HHS/United States
- AG019391/AG/NIA NIH HHS/United States
- R01 AG025440-04/AG/NIA NIH HHS/United States
- R01 AG019391-09/AG/NIA NIH HHS/United States
- R01 AG025440/AG/NIA NIH HHS/United States
- AG144051/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous