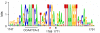RNA-directed DNA methylation induces transcriptional activation in plants - PubMed (original) (raw)
RNA-directed DNA methylation induces transcriptional activation in plants
Kenichi Shibuya et al. Proc Natl Acad Sci U S A. 2009.
Abstract
A class-C floral homeotic gene of Petunia, pMADS3, is specifically expressed in the stamen and carpels of developing flowers. We had previously reported the ect-pMADS3 phenomenon in which introduction of a part of the pMADS3 genomic sequence, including intron 2, induces ectopic expression of endogenous pMADS3. Unlike transcriptional or posttranscriptional gene silencing triggered by the introduction of homologous sequences, this observation is unique in that the gene expression is up-regulated. In this study, we demonstrated that the ect-pMADS3 phenomenon is due to transcriptional activation based on RNA-directed DNA methylation (RdDM) occurring in a particular CG in a putative cis-element in pMADS3 intron 2. The CG methylation was maintained over generations, along with pMADS3 ectopic expression, even in the absence of RNA triggers. These results demonstrate a previously undescribed transcriptional regulatory mechanism that could lead to the generation of a transcriptionally active epiallele, thereby contributing to plant evolution. Our results also reveal a putative negative cis-element for organ-specific transcriptional regulation of class-C floral homeotic genes, which could be difficult to identify by other approaches.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Targeting of pMADS3 intron 2 subregions by inverted repeat (IR) sequences. (A) Diagrams for target regions of IR expression in pMADS3 intron 2. Scales represent nucleotide numbers with the first nucleotide of pMADS3 intron 2 as nt 1. To the right are the numbers of transformant lines showing ect-pMADS3 (E), weak ect-pMADS3 (WE), pMADS3 silencing (S), and normal (N) phenotypes (9). (B) Flower phenotypes of transformant plants expressing IRs. (Bi) A flower of wild-type petunia (V26); (Bii) a typical flower of V002 plants showing the ect-pMADS3 phenotype with antheroid sectors along midveins and reduced petal limbs; flowers of V016 (Biii) and V019 (Biv) plants showing ect-pMADS3 phenotypes with antheroid tissues on petal limbs; (Bv) a flower of V019 plants showing strong ect-pMADS3 phenotype with antheroid petals (some flowers showed strong phenotypes as shown but others in the same plants showed milder ones); (Bvi) a typical flower of V034 plants showing strong ect-pMADS3 phenotype with petal-to-anther conversion; (Bvii) extreme flower phenotype of 71–14 × M (_T_1) showing complete conversion of petals to anthers; and (Bviii) curled cauline leaves of the V019 line. (C) Levels of pMADS3 transcripts in leaves (lf), sepals (se), petals (pl), stamens (st), and carpels (ca) of transgenic lines. Expression levels of pMADS3 relative to those of the elongation factor gene (EF) were measured by RT-qPCR. Mean ± SE (n = 3).
Fig. 2.
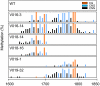
DNA methylation patterns in pMADS3 intron 2 of inverted repeat (IR)-expressed transgenic plants. V016 and V019 plants show the ect-MADS3 phenotype, whereas V018 plants show normal phenotypes. At least 9 clones are sequenced for each bisulfite-treated sample. Bars represent proportions of methylated clones at each site; red, blue, and black bars represent CG, CNG, and CNN sites, respectively. The scale represents nucleotide numbers in pMADS3 intron 2 with the first nucleotide of intron 2 as nt 1.
Fig. 3.
DNA methylation patterns in outcrossed pMADS3:GUS plants. (A) Inheritance of individual pMADS3 alleles in the _T_1 generation. Transgenes segregated out in both M × 73-6_9 and 71-14 × M_2 plants showing the ect-pMADS3 phenotype. (B) Allele-specific expression of endogenous pMADS3 in the petals of pMADS3:_GUS T_1 plants. The transformants analyzed were those without transgenes showing the ect-pMADS3 phenotype. The allele-specific expression was analyzed as described previously (9). M: marker; lane 1: 71-14 × M_2; lane 2: M × 73-6_9; lane 3: M × 73-6_7; lane 4: 71-14 × M_11. (C) DNA methylation patterns in _pMADS3_-mi and _pMADS3_-su2 alleles of M × 73-6_9 and 71-14 × M_2 plants. Bars represent proportions of methylated clones at each site; red, blue, and black bars represent CG, CNG, and CNN sites, respectively. The scale represents nucleotide numbers in pMADS3 intron 2 with the first nucleotide of intron 2 as nt 1.
Fig. 4.
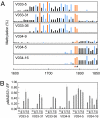
DNA methylation patterns and pMADS3 expression in V033 and V034 plants. (A) DNA methylation patterns in the 1600–1850 region. Bars represent proportions of methylated clones at each site; red, blue, and black bars represent CG, CNG, and CNN sites, respectively. The scale represents nucleotide numbers in pMADS3 intron 2 with the first nucleotide of intron 2 as nt 1. Asterisks indicate CG sites at nts 1768 (red) and 1771 (black). (B) pMADS3 mRNA levels in leaves (lf), sepals (se), petals (pl), stamens (st), and carpels (ca) of transgenic lines. Expression levels of pMADS3 relative to those of the elongation factor gene (EF) were measured by RT-qPCR. Mean ± SE (n = 3).
Fig. 5.
Sequence logo for conserved motifs in the intron 2 of pMADS3 homologs. The sequence logo (created by weblogo.berkeley.edu) was generated from 15 pMADS3 homologs of 13 plant species belonging to 9 different families (see
Table S1
). Asterisks indicate CG sites at nts 1768 (red) and 1771 (black). Numbers at the bottom indicate the positions in pMADS3 intron 2.
Similar articles
- Transgene-triggered, epigenetically regulated ectopic expression of a flower homeotic gene pMADS3 in Petunia.
Kapoor M, Baba A, Kubo K, Shibuya K, Matsui K, Tanaka Y, Takatsuji H. Kapoor M, et al. Plant J. 2005 Sep;43(5):649-61. doi: 10.1111/j.1365-313X.2005.02481.x. Plant J. 2005. PMID: 16115063 - Role of petunia pMADS3 in determination of floral organ and meristem identity, as revealed by its loss of function.
Kapoor M, Tsuda S, Tanaka Y, Mayama T, Okuyama Y, Tsuchimoto S, Takatsuji H. Kapoor M, et al. Plant J. 2002 Oct;32(1):115-27. doi: 10.1046/j.1365-313x.2002.01402.x. Plant J. 2002. PMID: 12366805 - Ectopic expression of pMADS3 in transgenic petunia phenocopies the petunia blind mutant.
Tsuchimoto S, van der Krol AR, Chua NH. Tsuchimoto S, et al. Plant Cell. 1993 Aug;5(8):843-53. doi: 10.1105/tpc.5.8.843. Plant Cell. 1993. PMID: 8104573 Free PMC article. - RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants.
Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJ, Matzke M. Huettel B, et al. Biochim Biophys Acta. 2007 May-Jun;1769(5-6):358-74. doi: 10.1016/j.bbaexp.2007.03.001. Epub 2007 Mar 12. Biochim Biophys Acta. 2007. PMID: 17449119 Review. - RNA-directed DNA methylation.
Wassenegger M. Wassenegger M. Plant Mol Biol. 2000 Jun;43(2-3):203-20. doi: 10.1023/a:1006479327881. Plant Mol Biol. 2000. PMID: 10999405 Review.
Cited by
- Epigenetic Dynamics and Regulation of Plant Male Reproduction.
Hou Q, Zhang T, Qi Y, Dong Z, Wan X. Hou Q, et al. Int J Mol Sci. 2022 Sep 9;23(18):10420. doi: 10.3390/ijms231810420. Int J Mol Sci. 2022. PMID: 36142333 Free PMC article. Review. - Omics Potential in Herbicide-Resistant Weed Management.
Patterson EL, Saski C, Küpper A, Beffa R, Gaines TA. Patterson EL, et al. Plants (Basel). 2019 Dec 14;8(12):607. doi: 10.3390/plants8120607. Plants (Basel). 2019. PMID: 31847327 Free PMC article. Review. - Epigenetic regulation of plant immunity: from chromatin codes to plant disease resistance.
Xie SS, Duan CG. Xie SS, et al. aBIOTECH. 2023 Mar 17;4(2):124-139. doi: 10.1007/s42994-023-00101-z. eCollection 2023 Jun. aBIOTECH. 2023. PMID: 37581024 Free PMC article. Review. - RNA-directed DNA methylation and demethylation in plants.
Chinnusamy V, Zhu JK. Chinnusamy V, et al. Sci China C Life Sci. 2009 Apr;52(4):331-43. doi: 10.1007/s11427-009-0052-1. Epub 2009 Apr 21. Sci China C Life Sci. 2009. PMID: 19381459 Free PMC article. Review. - Promoting gene expression in plants by permissive histone lysine methylation.
Cazzonelli CI, Millar T, Finnegan EJ, Pogson BJ. Cazzonelli CI, et al. Plant Signal Behav. 2009 Jun;4(6):484-8. doi: 10.4161/psb.4.6.8316. Epub 2009 Jun 2. Plant Signal Behav. 2009. PMID: 19816124 Free PMC article. Review.
References
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. - PubMed
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. - PubMed
- Alleman M, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources