The peopling of the Pacific from a bacterial perspective - PubMed (original) (raw)
. 2009 Jan 23;323(5913):527-30.
doi: 10.1126/science.1166083.
Bodo Linz, Yoshio Yamaoka, Helen M Windsor, Sebastien Breurec, Jeng-Yih Wu, Ayas Maady, Steffie Bernhöft, Jean-Michel Thiberge, Suparat Phuanukoonnon, Gangolf Jobb, Peter Siba, David Y Graham, Barry J Marshall, Mark Achtman
Affiliations
- PMID: 19164753
- PMCID: PMC2827536
- DOI: 10.1126/science.1166083
The peopling of the Pacific from a bacterial perspective
Yoshan Moodley et al. Science. 2009.
Abstract
Two prehistoric migrations peopled the Pacific. One reached New Guinea and Australia, and a second, more recent, migration extended through Melanesia and from there to the Polynesian islands. These migrations were accompanied by two distinct populations of the specific human pathogen Helicobacter pylori, called hpSahul and hspMaori, respectively. hpSahul split from Asian populations of H. pylori 31,000 to 37,000 years ago, in concordance with archaeological history. The hpSahul populations in New Guinea and Australia have diverged sufficiently to indicate that they have remained isolated for the past 23,000 to 32,000 years. The second human expansion from Taiwan 5000 years ago dispersed one of several subgroups of the Austronesian language family along with one of several hspMaori clades into Melanesia and Polynesia, where both language and parasite have continued to diverge.
Figures
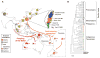
Fig. 1
(A) The distribution of H. pylori populations in Asia and the Pacific. The proportions of haplotypes at each sampling location (red numbers; table S1) that were assigned to different bacterial populations are displayed as pie charts whose sizes indicate the numbers of haplotypes. The geographic location of Melanesia and Polynesia is depicted. The term “Austronesia” refers to the entire region inhabited by Austronesian-speaking people from Madagascar through to the Easter Islands. (Inset) A detailed map of Taiwan showing the distribution of aboriginal tribes. The names of the tribes plus the proportion of hspMaori haplotypes among all haplotypes are shown in black at the right. The language-family designations are the same as the tribal names except where indicated by parentheses (EF, East Formosan; MP, Malayo-Polynesian). (B) Phylogenetic relationships among hspMaori strains (80% consensus of 100 ClonalFrame analyses). One haplotype each of hpAsia2 and hspEAsia was used to root the tree. Strains are color-coded according to Austronesian language family in (A). Two black circles within the Pacific clade indicate haplotypes isolated from the Torres Strait islands, and a black triangle among indigenous Taiwanese indicates an hspMaori haplotype from Yami.
Fig. 2
Global patterns of migration between eight pairs of H. pylori populations as calculated by the isolation with migration model (IMa). (A) Map. The magnitudes of migration are denoted by numbers and arrow thickness and their direction is indicated in blue or red. (B) Graph showing a linear relation between the calibration time (table S2) of six events (filled blue circles) that are dated by archaeological estimates and the estimated time (t). (C) Population tree reconstructed from a consensus of 1000 bootstrap samples from the range of calculated t values to determine the ages of nodes (thousands of years, kyr) associated with the peopling of the Sahul (unfilled circles). Ages (in light blue) are the 95% confidence limits of estimated coalescence times obtained by applying global rate minimum deformation (GRMD) rate-smoothing, as implemented in Treefinder, to the range of t values within the limits of calibration dates.
Fig. 3
Global phylogeny of H. pylori as calculated by a haplotype approach based on the 80% consensus of 100 ClonalFrame analyses. (A) Phylogenetic tree of divergence time, as indicated by node height versus geographic sources (bottom line) and population assignments (second line). Detailed sources of clades within populations are indicated in the third line from the bottom. Node heights were used to date the two hpSahul nodes (unfilled circles) based on six calibration times (filled blue circles, table S2). Age ranges (light blue numbers) are the 95% confidence limits of estimated coalescence times obtained with GRMD rate-smoothing over the range of node height values and calibration time limits. hpAFR2, hpAfrica2; hpAFR1, hpAfrica1; AM, America. (B) Graph showing a linear relation of calibration time with the range of heights for each node.
Comment in
- Anthropology. Where bacteria and languages concur.
Renfrew C. Renfrew C. Science. 2009 Jan 23;323(5913):467-8. doi: 10.1126/science.1168953. Science. 2009. PMID: 19164735 No abstract available.
Similar articles
- The impact of the Austronesian expansion: evidence from mtDNA and Y chromosome diversity in the Admiralty Islands of Melanesia.
Kayser M, Choi Y, van Oven M, Mona S, Brauer S, Trent RJ, Suarkia D, Schiefenhövel W, Stoneking M. Kayser M, et al. Mol Biol Evol. 2008 Jul;25(7):1362-74. doi: 10.1093/molbev/msn078. Epub 2008 Apr 3. Mol Biol Evol. 2008. PMID: 18390477 - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Using Experience Sampling Methodology to Capture Disclosure Opportunities for Autistic Adults.
Love AMA, Edwards C, Cai RY, Gibbs V. Love AMA, et al. Autism Adulthood. 2023 Dec 1;5(4):389-400. doi: 10.1089/aut.2022.0090. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116059 Free PMC article. - Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.
Pillay J, Gaudet LA, Saba S, Vandermeer B, Ashiq AR, Wingert A, Hartling L. Pillay J, et al. Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article. - Gestational Diabetes Mellitus: Unveiling Maternal Health Dynamics from Pregnancy Through Postpartum Perspectives.
Mora-Ortiz M, Rivas-García L. Mora-Ortiz M, et al. Open Res Eur. 2024 Nov 12;4:164. doi: 10.12688/openreseurope.18026.1. eCollection 2024. Open Res Eur. 2024. PMID: 39355538 Free PMC article. Review.
Cited by
- Genome-wide insights into the genetic history of human populations.
Pugach I, Stoneking M. Pugach I, et al. Investig Genet. 2015 Apr 1;6:6. doi: 10.1186/s13323-015-0024-0. eCollection 2015. Investig Genet. 2015. PMID: 25834724 Free PMC article. - Genetic populations and virulence factors of Helicobacter pylori.
Kabamba ET, Tuan VP, Yamaoka Y. Kabamba ET, et al. Infect Genet Evol. 2018 Jun;60:109-116. doi: 10.1016/j.meegid.2018.02.022. Epub 2018 Feb 19. Infect Genet Evol. 2018. PMID: 29471116 Free PMC article. Review. - Evolutionary history of Helicobacter pylori sequences reflect past human migrations in Southeast Asia.
Breurec S, Guillard B, Hem S, Brisse S, Dieye FB, Huerre M, Oung C, Raymond J, Tan TS, Thiberge JM, Vong S, Monchy D, Linz B. Breurec S, et al. PLoS One. 2011;6(7):e22058. doi: 10.1371/journal.pone.0022058. Epub 2011 Jul 19. PLoS One. 2011. PMID: 21818291 Free PMC article. - Population genetic structure and isolation by distance of Helicobacter pylori in Senegal and Madagascar.
Linz B, Vololonantenainab CR, Seck A, Carod JF, Dia D, Garin B, Ramanampamonjy RM, Thiberge JM, Raymond J, Breurec S. Linz B, et al. PLoS One. 2014 Jan 30;9(1):e87355. doi: 10.1371/journal.pone.0087355. eCollection 2014. PLoS One. 2014. PMID: 24498084 Free PMC article. - Transmission of the PabI family of restriction DNA glycosylase genes: mobility and long-term inheritance.
Kojima KK, Kobayashi I. Kojima KK, et al. BMC Genomics. 2015 Oct 19;16:817. doi: 10.1186/s12864-015-2021-3. BMC Genomics. 2015. PMID: 26481899 Free PMC article.
References
- Macaulay V, et al. Science. 2005;308:1034. - PubMed
- Pope KO, Terrell JE. J Biogeography. 2008;35:1.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous