Reuse, replace, recycle. Specificity in subunit inheritance and assembly of higher-order septin structures during mitotic and meiotic division in budding yeast - PubMed (original) (raw)
Review
Reuse, replace, recycle. Specificity in subunit inheritance and assembly of higher-order septin structures during mitotic and meiotic division in budding yeast
Michael A McMurray et al. Cell Cycle. 2009.
Abstract
Septins are guanine nucleotide-binding proteins that form hetero-oligomeric complexes, which assemble into filaments and higher-order structures at sites of cell division and morphogenesis in eukaryotes. Dynamic changes in the organization of septin-containing structures occur concomitantly with progression through the mitotic cell cycle and during cell differentiation. Septins also undergo stage-specific post-translational modifications, which have been implicated in regulating their dynamics, in some cases via purported effects on septin turnover. In our recent study, the fate of two of the five septins expressed in mitotic cells of budding yeast (Saccharomyces cerevisiae) was tracked using two complementary fluorescence-based methods for pulse-chase analysis. During mitotic growth, previously-made molecules of both septins (Cdc10 and Cdc12) persisted through multiple successive divisions and were incorporated equivalently with newly synthesized molecules into hetero-oligomers and higher-order structures. Similarly, in cells undergoing meiosis and the developmental program of sporulation, pre-existing copies of Cdc10 were incorporated into new structures. In marked contrast, Cdc12 was irreversibly excluded from septin complexes and replaced by another septin, Spr3. Here, we discuss the broader implications of these results and related findings with regard to how septin dynamics is coordinated with the mitotic cell cycle and in the yeast life cycle, and how these observations may relate to control of the dynamics of other complex multi-subunit assemblies.
Figures
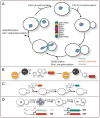
Figure 1
Recycling of budding yeast septin subunits during the mitotic cell division cycle revealed by fluorescent pulse-chase analysis. (A) The mitotic cell division cycle of budding yeast is schematized with the location of septins (green), spindle pole bodies (red), and the nucleus (blue) indicated as landmarks of cell cycle progression. Also indicated are the approximate stages when post-translational modifications are added to septins and, where known, when removed. Center, arrangement of individual septin subunits within the hetero-octameric rod. (B) The SNAP-tag™, a domain (black) of the human O-alkyl-guanine-DNA alkyltransferase (hAGT), shown fused in-frame to the C terminus of the yeast septin Cdc12 (orange), can be covalently labeled in vivo by reaction of its active site Cys with a benzyl guanine (BG) derivatized with a fluor (red). (C) Use of the SNAP-tag for one-color-labeling for pulse-chase analysis and for two-color labeling to monitor potential age-related differences. For pulse-chase analysis, cells expressing a SNAP-tagged septin are incubated for a short time in medium containing a cell-permeable fluorescently labeled BG derivative (the “pulse”), the cells are then washed to remove any unreacted dye, and the fate of the population of labeled protein (red) is monitored under the fluorescence microscope over the course of multiple subsequent cell divisions (the “chase”). For two-color labeling to compare the behavior of older and newer molecules, the cells are allowed to propagate for only 2–3 divisions after the first labeling, which provides sufficient time for synthesis of new unlabeled molecules without reducing too drastically the level of the previously labeled molecules. The new molecules can then be labeled by incubating the cells in medium containing a second BG derivative containing a fluor of a different color (green). (D) Use of differential gene expression for two-color labeling to monitor potential age-related differences. Expression of Cdc10-GFP from the HO promoter (“_PHO_”) is repressed in diploid cells, whereas expression of Cdc10-mCherry from the CDC10 promoter (“_PCDC10_”) is constitutive. Following meiosis and sporulation, the HO promoter only becomes active after the haploid spore has undergone its first mitotic division and, hence, Cdc10-GFP is produced during the second mitotic division.
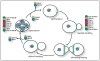
Figure 2
Certain septin subunits are recycled, whereas others are discarded, during developmental transitions in the budding yeast life cycle. Localization of Cdc10 (green) is also representative of the localization of Cdc3 and Cdc11. Septins form a broad and very diffuse band at the base of the mating projection, or shmoo. Depiction of the organization of the septin subunits within the hetero-octamers present in sporulating cells is inferred from biochemical and genetic evidence, but has not been confirmed by methods for ultrastructural analysis. It is not known whether Cdc12 and Shs1 are free or remain associated with each other when they are excluded from septin hetero-octamers during sporulation, or whether Spr3 and Spr28 are free or remain associated with each other when they are displaced from hetero-octamers during the resumption of mitotic growth.
Figure 3
Alternative fates of a septin subunit in budding yeast. A generic septin subunit is followed diagrammatically from its initial translation and early folding steps (“new synthesis”) through subsequent incorporation with other subunits into hetero-octamers. Regulatory modifications directing assembly into higher-order structures are presumably exerted on the hetero-octamer and their removal may promote disassembly (or other active modifications may do so). In any event, upon disassembly, all such modifications must be erased or reset to allow recycling of pre-made hetero-octamers into higher-order structures in the next cell cycle. Replacement of damaged subunits or substitution of developmentally-specific subunits presumably involves partial disassembly of hetero-octamers and the incorporation of new subunits. Whether disassembly can proceed to the level of free monomers is not known. During developmental transitions, when certain subunits are excluded from hetero-octamers and replaced by other specialized subunits, this exclusion is not accompanied by degradation (and whether the excluded subunit is monomeric or in complex with other excluded subunits remains unknown). The cytosolic chaperonin complex CCT (also called TRiC) may be involved in the folding of individual septin subunits and/or in the assembly of subunits into hetero-octamers and/or in the exchange of subunits into and out of hetero-octamers. GTP binding (not shown) has an important role in stabilizing the tertiary structure of each septin and in promoting stable hetero-octamer assembly, but genetic and biochemical evidence indicates that active cycles of GTP binding and hydrolysis by septins play a minimal, if any, role in septin hetero-octamer dynamics and in the formation of higher-order structures.
Similar articles
- Septin stability and recycling during dynamic structural transitions in cell division and development.
McMurray MA, Thorner J. McMurray MA, et al. Curr Biol. 2008 Aug 26;18(16):1203-8. doi: 10.1016/j.cub.2008.07.020. Epub 2008 Aug 14. Curr Biol. 2008. PMID: 18701287 Free PMC article. - Guanidine hydrochloride reactivates an ancient septin hetero-oligomer assembly pathway in budding yeast.
Johnson CR, Steingesser MG, Weems AD, Khan A, Gladfelter A, Bertin A, McMurray MA. Johnson CR, et al. Elife. 2020 Jan 28;9:e54355. doi: 10.7554/eLife.54355. Elife. 2020. PMID: 31990274 Free PMC article. - Assembly, molecular organization, and membrane-binding properties of development-specific septins.
Garcia G 3rd, Finnigan GC, Heasley LR, Sterling SM, Aggarwal A, Pearson CG, Nogales E, McMurray MA, Thorner J. Garcia G 3rd, et al. J Cell Biol. 2016 Feb 29;212(5):515-29. doi: 10.1083/jcb.201511029. J Cell Biol. 2016. PMID: 26929450 Free PMC article. - Here come the septins: novel polymers that coordinate intracellular functions and organization.
Spiliotis ET, Nelson WJ. Spiliotis ET, et al. J Cell Sci. 2006 Jan 1;119(Pt 1):4-10. doi: 10.1242/jcs.02746. J Cell Sci. 2006. PMID: 16371649 Free PMC article. Review. - Some assembly required: yeast septins provide the instruction manual.
Versele M, Thorner J. Versele M, et al. Trends Cell Biol. 2005 Aug;15(8):414-24. doi: 10.1016/j.tcb.2005.06.007. Trends Cell Biol. 2005. PMID: 16009555 Free PMC article. Review.
Cited by
- A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance.
Babour A, Bicknell AA, Tourtellotte J, Niwa M. Babour A, et al. Cell. 2010 Jul 23;142(2):256-69. doi: 10.1016/j.cell.2010.06.006. Epub 2010 Jul 8. Cell. 2010. PMID: 20619447 Free PMC article. - GTP-induced conformational changes in septins and implications for function.
Sirajuddin M, Farkasovsky M, Zent E, Wittinghofer A. Sirajuddin M, et al. Proc Natl Acad Sci U S A. 2009 Sep 29;106(39):16592-7. doi: 10.1073/pnas.0902858106. Epub 2009 Sep 15. Proc Natl Acad Sci U S A. 2009. PMID: 19805342 Free PMC article. - Microtubules support a disk-like septin arrangement at the plasma membrane of mammalian cells.
Sellin ME, Holmfeldt P, Stenmark S, Gullberg M. Sellin ME, et al. Mol Biol Cell. 2011 Dec;22(23):4588-601. doi: 10.1091/mbc.E11-09-0754. Epub 2011 Oct 12. Mol Biol Cell. 2011. PMID: 21998205 Free PMC article. - Septin filament formation is essential in budding yeast.
McMurray MA, Bertin A, Garcia G 3rd, Lam L, Nogales E, Thorner J. McMurray MA, et al. Dev Cell. 2011 Apr 19;20(4):540-9. doi: 10.1016/j.devcel.2011.02.004. Dev Cell. 2011. PMID: 21497764 Free PMC article. - The MoLfa1 Protein Regulates Fungal Development and Septin Ring Formation in Magnaporthe oryzae.
Wu JQ, Zhu XM, Bao JD, Wang JY, Yu XP, Lin FC, Li L. Wu JQ, et al. Int J Mol Sci. 2024 Mar 19;25(6):3434. doi: 10.3390/ijms25063434. Int J Mol Sci. 2024. PMID: 38542408 Free PMC article.
References
- Farkasovsky M, Herter P, Voss B, Wittinghofer A. Nucleotide binding and filament assembly of recombinant yeast septin complexes. Biol Chem. 2005;386:643–56. - PubMed
- Vrabioiu AM, Gerber SA, Gygi SP, Field CM, Mitchison TJ. The majority of the Saccharomyces cerevisiae septin complexes do not exchange guanine nucleotides. J Biol Chem. 2004;279:3111–8. - PubMed
- Hartwell LH. Genetic control of the cell division cycle in yeast IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K99 GM086603/GM/NIGMS NIH HHS/United States
- R01 GM021841/GM/NIGMS NIH HHS/United States
- R01 GM101314/GM/NIGMS NIH HHS/United States
- R01 GM21841/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases