Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA - PubMed (original) (raw)
Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA
Jun Katahira et al. EMBO J. 2009.
Abstract
In metazoans, nuclear export of bulk mRNA is mediated by Tap-p15, a conserved heterodimeric export receptor that cooperates with adaptor RNA-binding proteins. In this article, we show that Thoc5, a subunit of the mammalian TREX complex, binds to a distinct surface on the middle (Ntf2-like) domain of Tap. Notably, adaptor protein Aly and Thoc5 can simultaneously bind to non-overlapping binding sites on Tap-p15. In vivo, Thoc5 was not required for bulk mRNA export. However, nuclear export of HSP70 mRNA depends on both Thoc5 and Aly. Consistent with a function as a specific export adaptor, Thoc5 exhibits in vitro RNA-binding activity and is associated with HSP70 mRNPs in vivo as a component of the stable THO complex. Thus, through the combinatorial use of an adaptor (e.g., Aly) and co-adapter (e.g., Thoc5), Tap-p15 could function as an export receptor for different classes of mRNAs.
Figures
Figure 1
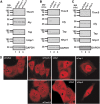
Thoc5 is a novel interaction counterpart of Tap. (A) Domain organization and the known interaction counterparts of the Tap-p15 heterodimer. Numbers on each rectangle indicate amino-acid positions of human Tap (for details, see Segref et al, 1997; Liker et al, 2000; Rodrigues et al, 2001; Reed and Hurt, 2002). (B) Crude lysates of E. coli with (indicated by +) or without (indicated by −) Thoc5–FLAG expression were added to GSH beads pre-adsorbed to GST or GST fused to various fragments of Tap. Bound proteins were analysed by SDS–PAGE followed by CBB staining (upper panel) and western blot using anti-FLAG antibody (lower panel). In lanes 13 and 14, aliquots of 10% of each input were loaded. Arrowheads indicate the positions of Thoc5–FLAG. Positions of molecular weight markers are indicated on the left in kDa. Note that in this particular gel, p15 migrated to the dye front. (C) Purified Thoc5–FLAG was added to GSH beads pre-adsorbed to GST (lane 1) or GST fused to various fragments of Tap (lanes 2–6). Aliquots of 25% of each bound fraction were separated by SDS—PAGE, and protein bands were detected by CBB staining (upper panel) and western blot using anti-FLAG antibody (lower panel). In lane 7, a total of 10% of input was loaded. Arrowheads indicate the positions of p15 and Thoc5–FLAG, whereas asterisks indicate the positions of each GST-fusion protein. Positions of molecular weight markers are indicated on the left in kDa. (D) Surface representation showing the Ntf2-like domain of Tap (blue) complexed with p15 (magenta) and FG-containing peptide (orange) (Fribourg et al, 2001). Different surfaces are also viewed from orientations indicated by the arrows in the upper-most figure. The regions of Tap that are critical for Thoc5 binding are coloured in yellow. An arrowhead indicates the short loop (aa 505–507) of Tap. The positions of alanine-scan mutations are indicated at the bottom of the figures as a single-letter code. The numbers on top of the sequence indicate the amino-acid positions of Tap. The residues shaded in yellow correspond to the region coloured in yellow in the 3D model. (E) Same as in (C), but Tap (188–619) containing the alanine-scan mutations complexed with p15 were used. Arrowheads show the positions of p15 and Thoc5–FLAG on the gel. Positions of molecular weight markers are indicated on the left in kDa. (F) Full-length and different domains (aa 1–251 and 251–683) of Thoc5 were expressed in E. coli as His6- and FLAG double-tagged proteins. Purified proteins (5% of input, lanes 7–9) were pulled down by GST (lanes 1–3) or GST-Tap (188-619)-p15 (lanes 4–6). Proteins in bound fractions (25%) were analysed by SDS–PAGE followed by CBB staining (upper) and western blot using anti-FLAG antibody (lower). The positions of Thoc5–FLAG and its derivative are indicated by asterisks, whereas those of Tap (188–619), GST and p15 are indicated by arrowheads. Positions of molecular weight markers are shown on the left in kDa.
Figure 2
Thoc5 shuttles between the nucleus and the cytoplasm. (A) Total cell extracts prepared from parental L929 cells (lane 1) or L929 cell lines stably expressing Thoc5–GFP (lane 2) and hHpr1–GFP (lane 3) were subjected to western blot using anti-GFP antibody. Positions of molecular weight markers are indicated on the left in kDa. (B, C) Heterokaryon formation was performed using Xenopus A6 cells and the L929 cell lines stably expressing Thoc5–GFP (B) or hHpr1–GFP (C) in the presence of CHX. After incubation for 3 h at 30°C in the presence of CHX, the cells were fixed and immunostained with anti-Tap (shuttling; upper panels) and anti-hnRNP C (non-shuttling; lower panels) antibodies. Nuclei were stained with Hoechst 33342 dye. Arrows indicate Xenopus nuclei in fused cells, whereas arrowheads indicate those in unfused cells. Insets show magnified views of the GFP signals in the fused cells.
Figure 3
Depletion of Thoc5 does not affect bulk poly(A)+ RNA export in mammalian cells. (A) HeLa cells were treated with the indicated siRNAs for 72 h. Total cell extracts were subjected to western blot using the indicated antibodies. For a negative control, siRNA against DsRed protein was used. Positions of molecular weight markers are indicated on the left in kDa. (B) HeLa cells were treated with the indicated siRNAs for 72 h. Total cell extracts were subjected to western blot using the indicated antibodies. For a negative control, siRNA against DsRed protein was used. Positions of molecular weight markers are indicated on the left in kDa. (C) HeLa cells were treated with the indicated siRNAs for 45 h. Total cell extracts prepared from the cells were subjected to western blot using the indicated antibodies. For a negative control, siRNA against DsRed protein was used. Positions of molecular weight markers are indicated on the left in kDa. (D) HeLa cells grown on glass-bottomed dishes were treated with the indicated siRNAs for either 45 h (siRNAs against Tap) or 72 h (others). The cells were fixed and subjected to in situ hybridization using Cy3-labelled oligo-dT50 probe.
Figure 4
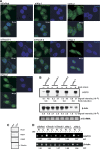
HSP70 mRNA export requires both Thoc5 and Aly. (A) siRNAs against DsRed (siDsRed: negative control), Aly (siAly-1 and -2), Thoc5 (siThoc5-1 and -2) or Tap (siTap-1 and -2) were transfected to HeLa cells. At 48 h (for siTap-1 and -2) or 72 h (for other siRNAs) post-transfection, the cells were subjected to heat shock at 43°C for 1 h, and in situ hybridization using FITC-labelled human HSP70 oligonucleotide probes was performed. (B) HeLa cells treated with indicated siRNAs for 72 h were left untreated (indicated by −) or temperature shifted at 43°C for 1 h (indicated by +). Total RNAs (5 μg per lane) prepared from the cells were subjected to northern blot analysis using [32P]-labelled HSP70 and β-actin cDNA probes. 28 S rRNA detected by ethidium bromide staining served as loading control. (C) HeLa cells were fractionated into nuclear (left) and cytoplasmic (right) fractions. Each fraction was subjected to SDS–PAGE followed by western blot using anti-RCC1 and anti-hHpr1 (nuclear markers) or anti-G3BP and anti-α-tubulin (cytoplasmic markers) antibodies. (D) HeLa cells treated with the indicated siRNAs for 72 h were temperature-shifted at 43°C for 1 h. Total RNA was isolated from nuclear (N) and cytoplasmic (C) fractions separated as in (C). HSP70 and β-actin mRNAs in each fraction was detected by RT–PCR using specific primers. Relative nuclear signals (N/C) are shown at the bottom of the panels.
Figure 5
Tap-p15, Thoc5 and Aly constitute a hetero-tetramer. (A) Purified Thoc5–FLAG (25 μg) along with increasing amounts of full-length Tap-p15 (lanes 2–4; 0, 8, 16 μg) was incubated with GSH-beads pre-adsorbed to purified GST-Aly (10 μg). In lane 5, Tap (188–619)-p15 (20 μg) was used instead of full-length Tap-p15. The beads were washed and aliquots of the bound fractions (25%) were analysed by SDS–PAGE followed by CBB staining. In lane 1, buffer alone was added in binding reaction for comparison. In lanes 6–8, purified Thoc5–FLAG (1.5 μg), Tap-p15 (2 μg) and Tap (188–619)-p15 (2.5 μg) were run as markers. The bound fractions (10%) were also subjected to western blot using the indicated antibodies. Positions of molecular weight markers are indicated on the left in kDa. (B) Purified Thoc5–FLAG (lanes 3, 5, 7) or buffer alone (lanes 2, 4, 6) was incubated with GSH-beads pre-adsorbed to GST (lanes 2 and 3), GST–Aly (lanes 4 and 5) and GST–hHpr1 (lanes 6 and 7). The beads were washed and aliquots of the bound fractions (25%) were analysed by SDS–PAGE followed by CBB staining and western blot using anti-FLAG antibody. In lane 1, 10% of input was loaded.
Figure 6
Thoc5 and Aly are components of the HSP70 mRNP. (A) RNA-binding assay was performed using a 91 bp [32P]-labelled RNA encoding pBluescript SK polylinker sequence. Increasing amounts of purified recombinant GST (lanes 2–4: 160, 320, 480 pmol), Thoc5–FLAG (lanes 5–8: 2, 4, 12, 20 pmol) and GST–Aly (lanes 9–12: 0.3, 0.6, 2, 3 pmol) proteins were added to total 10 μl of each binding reaction. Probe alone was run in lane 1. The binding reactions were separated by 6% polyacrylamide gel electrophoresis and visualized by autoradiography. A bracket indicates the positions of the protein–RNA complexes. (B) Competition assay of the RNA-binding activity of Thoc5 was performed using increasing amounts (10, 25, 100, 300 ng each) of ribo-homopolymers (lanes 3–18) and a dsDNA (lanes 19–22) as competitors. (C) Heat-shocked HeLa cells were subjected to RNA co-immunoprecipitation assay. Co-precipitated RNAs were then subjected to PCR using the indicated primer pairs. RNA isolated from the crude cell extract was also subjected to RT–PCR (lanes 1, 2, 10 and 11; input). In lanes indicated by RT, reverse transcriptase was omitted from the first-strand synthesis reactions. Amplified cDNA fragments were separated by 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining. DNA size markers were run on lane 9 (indicated by M) and the position of the 500 bp marker is indicated on the left. (D) Heat-shocked HeLa cells were subjected to RNA co-immunoprecipitation assay using the indicated antibodies as in (C). (E) Heat-shocked HeLa cells were subjected to RNA co-immunoprecipitation assay using anti-Thoc5 antibody. mRNPs released from the anti-Thoc5 beads were re-immunoprecipitated using the indicated antibodies. Co-precipitated RNAs were analysed as in (C).
Figure 7
Depletion of Thoc6 also blocks nuclear export of HSP70 mRNA. (A) HeLa cells were treated with the indicated siRNAs for 72 h. Total cell extracts prepared from the cells were subjected to western blot using the indicated antibodies. For a negative control, siRNA against DsRed protein was used. Positions of molecular weight markers are indicated on the left in kDa. (B) HeLa cells grown on glass-bottomed dishes were treated with the indicated siRNAs for 72 h. The cells were fixed and subjected to in situ hybridization using Cy3-labelled oligo-dT50 probe. (C) HeLa cells grown on glass-bottomed dishes were treated with the indicated siRNAs against Thoc6 for 72 h. The cells were fixed and subjected to in situ hybridization using the FITC-labelled HSP70 oligonucleotide probes. Cells treated with siThoc5-1 (upper right panel) are shown for comparison of the size of the nuclear foci.
Similar articles
- Identification of mRNAs that are spliced but not exported to the cytoplasm in the absence of THOC5 in mouse embryo fibroblasts.
Guria A, Tran DD, Ramachandran S, Koch A, El Bounkari O, Dutta P, Hauser H, Tamura T. Guria A, et al. RNA. 2011 Jun;17(6):1048-56. doi: 10.1261/rna.2607011. Epub 2011 Apr 27. RNA. 2011. PMID: 21525145 Free PMC article. - ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export.
Chen IH, Li L, Silva L, Sandri-Goldin RM. Chen IH, et al. J Virol. 2005 Apr;79(7):3949-61. doi: 10.1128/JVI.79.7.3949-3961.2005. J Virol. 2005. PMID: 15767397 Free PMC article. - Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA.
Chi B, Wang Q, Wu G, Tan M, Wang L, Shi M, Chang X, Cheng H. Chi B, et al. Nucleic Acids Res. 2013 Jan;41(2):1294-306. doi: 10.1093/nar/gks1188. Epub 2012 Dec 7. Nucleic Acids Res. 2013. PMID: 23222130 Free PMC article. - THOC5, a member of the mRNA export complex: a novel link between mRNA export machinery and signal transduction pathways in cell proliferation and differentiation.
Tran DD, Koch A, Tamura T. Tran DD, et al. Cell Commun Signal. 2014 Jan 10;12:3. doi: 10.1186/1478-811X-12-3. Cell Commun Signal. 2014. PMID: 24410813 Free PMC article. Review. - [Regulation of nuclear export and cytoplasmic localization of mRNAs by NXF family proteins].
Katahira J. Katahira J. Tanpakushitsu Kakusan Koso. 2009 Dec;54(16 Suppl):2109-13. Tanpakushitsu Kakusan Koso. 2009. PMID: 21089626 Review. Japanese. No abstract available.
Cited by
- Structural differences between the closely related RNA helicases, UAP56 and URH49, fashion distinct functional apo-complexes.
Fujita KI, Ito M, Irie M, Harada K, Fujiwara N, Ikeda Y, Yoshioka H, Yamazaki T, Kojima M, Mikami B, Mayeda A, Masuda S. Fujita KI, et al. Nat Commun. 2024 Jan 15;15(1):455. doi: 10.1038/s41467-023-44217-8. Nat Commun. 2024. PMID: 38225262 Free PMC article. - The Mammalian Ecdysoneless Protein Interacts with RNA Helicase DDX39A To Regulate Nuclear mRNA Export.
Saleem I, Mirza S, Sarkar A, Raza M, Mohapatra B, Mushtaq I, Kim JH, Mishra NK, Alsaleem MA, Rakha EA, Qiu F, Guda C, Band H, Band V. Saleem I, et al. Mol Cell Biol. 2021 Jun 23;41(7):e0010321. doi: 10.1128/MCB.00103-21. Epub 2021 Jun 23. Mol Cell Biol. 2021. PMID: 33941617 Free PMC article. - Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress.
Lv DW, Subburaj S, Cao M, Yan X, Li X, Appels R, Sun DF, Ma W, Yan YM. Lv DW, et al. Mol Cell Proteomics. 2014 Feb;13(2):632-52. doi: 10.1074/mcp.M113.030171. Epub 2013 Dec 11. Mol Cell Proteomics. 2014. PMID: 24335353 Free PMC article. - Messenger RNA export from the nucleus: a series of molecular wardrobe changes.
Kelly SM, Corbett AH. Kelly SM, et al. Traffic. 2009 Sep;10(9):1199-208. doi: 10.1111/j.1600-0854.2009.00944.x. Epub 2009 Jun 27. Traffic. 2009. PMID: 19552647 Free PMC article. Review. - A Sub-Element in PRE enhances nuclear export of intronless mRNAs by recruiting the TREX complex via ZC3H18.
Chi B, Wang K, Du Y, Gui B, Chang X, Wang L, Fan J, Chen S, Wu X, Li G, Cheng H. Chi B, et al. Nucleic Acids Res. 2014 Jun;42(11):7305-18. doi: 10.1093/nar/gku350. Epub 2014 Apr 29. Nucleic Acids Res. 2014. PMID: 24782531 Free PMC article.
References
- Aguilera A (2005) Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr Opin Cell Biol 17: 242–250 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous