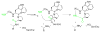Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates - PubMed (original) (raw)
Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates
Jaeheon Lee et al. J Am Chem Soc. 2009.
Abstract
Small N-C terminally cyclic ribosomal peptides are common in nature, yet the mechanisms underlying their biosyntheses remain largely unknown. We recently identified candidate peptide cyclase genes in the metagenomes of marine animals. Here we report that PatG, a protease discovered in this analysis, cleaves variables, short peptides out of a precursor protein, and cyclizes these peptides in vivo and in vitro.
Figures
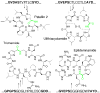
Figure 1
N-C cyclic peptides of the cyanobactin group. The partial precursor peptide sequences are shown that give rise to the cyclic products. Amide bonds formed by PatG and its relatives are shown in green, while recognition sequences are shown in bold and cassette sequences leading to products are underlined. Patellin 2, ulithiacyclamide, and trichamide are natural products that require multiple enzymatic processing steps in their synthesis, while the eptidemnamide precursor was engineered and requires only cleavage and cyclization.
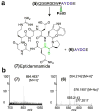
Figure 2. PatA catalyzes two N-terminal cleavage reactions
(a) The precursor peptide PatEdm is cleaved by PatA at a recognition sequence to release first (5) and then (4), with free N-termini for cyclization. (b) SDS-PAGE of a time-course experiment with PatA and PatEdm shows that the first site is slowly cleaved, followed by relatively rapid proteolysis of the second site. (c) All reaction products were observed by mass spectrometry
Figure 3. PatG catalyzes cyclization by a transamidation mechanism
(a) Starting from product (5) of PatA, PatG cleaves this peptide into a cyclic compound, eptidemnamide, and a small linear fragment representing the recognition sequence. (b) High resolution mass spectrometry and comparison to authentic standards confirmed that this cyclic peptide was formed from compound (5).
Figure 4. Proposed transamidation mechanism of peptide cyclization
In this simplified scheme, the active-site Ser of PatG is shown forming an enzyme-substrate tetrahedral intermediate with loss of the AYDGE recognition sequence. Subsequently, the N-terminus of the substrate attacks the activated ester bond to release a cyclic peptide and the free enzyme.
Similar articles
- Construction and screening of vast libraries of natural product-like macrocyclic peptides using in vitro display technologies.
Bashiruddin NK, Suga H. Bashiruddin NK, et al. Curr Opin Chem Biol. 2015 Feb;24:131-8. doi: 10.1016/j.cbpa.2014.11.011. Epub 2014 Dec 5. Curr Opin Chem Biol. 2015. PMID: 25483262 Review. - Macrocyclic Iminopeptides Diversify To Better Target Proteins.
Reguera L, Rivera DG. Reguera L, et al. ChemMedChem. 2020 Jul 3;15(13):1111-1112. doi: 10.1002/cmdc.202000261. Epub 2020 Jun 15. ChemMedChem. 2020. PMID: 32516501 Review. - Natural and Synthetic Macrocyclic Inhibitors of the Histone Deacetylase Enzymes.
Maolanon AR, Kristensen HM, Leman LJ, Ghadiri MR, Olsen CA. Maolanon AR, et al. Chembiochem. 2017 Jan 3;18(1):5-49. doi: 10.1002/cbic.201600519. Epub 2016 Dec 8. Chembiochem. 2017. PMID: 27748555 Review. - De Novo Discovery of Pseudo-Natural Prenylated Macrocyclic Peptide Ligands.
Inoue S, Thanh Nguyen D, Hamada K, Okuma R, Okada C, Okada M, Abe I, Sengoku T, Goto Y, Suga H. Inoue S, et al. Angew Chem Int Ed Engl. 2024 Sep 2;63(36):e202409973. doi: 10.1002/anie.202409973. Epub 2024 Jul 22. Angew Chem Int Ed Engl. 2024. PMID: 38837490 - Structures of cyanobactin maturation enzymes define a family of transamidating proteases.
Agarwal V, Pierce E, McIntosh J, Schmidt EW, Nair SK. Agarwal V, et al. Chem Biol. 2012 Nov 21;19(11):1411-22. doi: 10.1016/j.chembiol.2012.09.012. Chem Biol. 2012. PMID: 23177196 Free PMC article.
Cited by
- Cell-free biosynthesis and engineering of ribosomally synthesized lanthipeptides.
Liu WQ, Ji X, Ba F, Zhang Y, Xu H, Huang S, Zheng X, Liu Y, Ling S, Jewett MC, Li J. Liu WQ, et al. Nat Commun. 2024 May 21;15(1):4336. doi: 10.1038/s41467-024-48726-y. Nat Commun. 2024. PMID: 38773100 Free PMC article. - Cheminformatics-Guided Cell-Free Exploration of Peptide Natural Products.
Pelton JM, Hochuli JE, Sadecki PW, Katoh T, Suga H, Hicks LM, Muratov EN, Tropsha A, Bowers AA. Pelton JM, et al. J Am Chem Soc. 2024 Mar 27;146(12):8016-8030. doi: 10.1021/jacs.3c11306. Epub 2024 Mar 12. J Am Chem Soc. 2024. PMID: 38470819 Free PMC article. - Proteases Involved in Leader Peptide Removal during RiPP Biosynthesis.
Eslami SM, van der Donk WA. Eslami SM, et al. ACS Bio Med Chem Au. 2023 Dec 13;4(1):20-36. doi: 10.1021/acsbiomedchemau.3c00059. eCollection 2024 Feb 21. ACS Bio Med Chem Au. 2023. PMID: 38404746 Free PMC article. Review. - An Autocatalytic Peptide Cyclase Improves Fidelity and Yield of Circular Peptides In Vivo and In Vitro.
Lacerna N 2nd, Cong Y, Schmidt EW. Lacerna N 2nd, et al. ACS Synth Biol. 2024 Jan 19;13(1):394-401. doi: 10.1021/acssynbio.3c00645. Epub 2024 Jan 9. ACS Synth Biol. 2024. PMID: 38194299 Free PMC article. - Peptidase Activation by a Leader Peptide-Bound RiPP Recognition Element.
Kretsch AM, Gadgil MG, DiCaprio AJ, Barrett SE, Kille BL, Si Y, Zhu L, Mitchell DA. Kretsch AM, et al. Biochemistry. 2023 Feb 21;62(4):956-967. doi: 10.1021/acs.biochem.2c00700. Epub 2023 Feb 3. Biochemistry. 2023. PMID: 36734655 Free PMC article.
References
- Trabi M, Craik DJ. Trends Biochem Sci. 2002;27:132–138. - PubMed
- Scarborough RM. Am Heart J. 1999;138:1093–1104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM071425/GM/NIGMS NIH HHS/United States
- R01 GM071425/GM/NIGMS NIH HHS/United States
- R01 GM071425-01A1/GM/NIGMS NIH HHS/United States
- R01 GM071425-04/GM/NIGMS NIH HHS/United States
- R01 GM071425-03/GM/NIGMS NIH HHS/United States
- R01 GM071425-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases