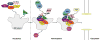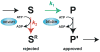Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines - PubMed (original) (raw)
Review
Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines
Jonathan P Staley et al. Curr Opin Cell Biol. 2009 Feb.
Abstract
Ribosomes and spliceosomes are ribonucleoprotein nanomachines that catalyze translation of mRNA to synthesize proteins and splicing of introns from pre-mRNAs, respectively. Assembly of ribosomes involves more than 300 proteins and RNAs, and that of spliceosomes over 100 proteins and RNAs. Construction of these enormous ribonucleoprotein particles (RNPs) is a dynamic process, in which the nascent RNPs undergo numerous ordered rearrangements of RNA-RNA, RNA-protein, and protein-protein interactions. Here we outline similar principles that have emerged from studies of ribosome and spliceosome assembly. Constituents of both RNPs form subassembly complexes, which can simplify the task of assembly and segregate functions of assembly factors. Reorganization of RNP topology, and proofreading of proper assembly, are catalyzed by protein- or RNA-dependent ATPases or GTPases. Dynamics of intermolecular interactions may be facilitated or regulated by cycles of post-translational modifications. Despite this repertoire of tools, mistakes occur in RNP assembly or in processing of RNA substrates. Quality control mechanisms recognize and turnover misassembled RNPs and reject improper substrates.
Figures
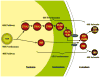
Fig. 1
Maturation of preribosomes in Saccharomyces cerevisiae. Ribosome biogenesis begins in the nucleolus, where pre-rRNA is transcribed and packaged into the 90S pre-rRNP, together with a subset of ribosomal proteins and ribosome assembly factors. Subsequent steps of maturation occur in the nucleolus, nucleoplasm and cytoplasm. The 90S pre-rRNP is converted into the 66S and 43S particles by cleavage within the pre-rRNA. There are at least six consecutive 66S precursors to mature 60S ribosomal subunits, distinguished by the consecutive pre-rRNA processing intermediates contained within them. The 43S pre-rRNP containing 20S pre-rRNA is exported to the cytoplasm where mature 40S subunits are formed.
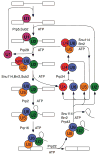
Fig. 2
The spliceosome cycle. The spliceosome assembles de novo on a pre-mRNA transcript, catalyzes intron removal, dissociates the products and disassembles to permit recycling for subsequent rounds of splicing. Numerous ATP-dependent steps require factors belonging to the DExD/H box family of proteins. Revised from Cell 1998 **92:**315–326, with permission from Elsevier.
Fig. 3
Coupling assembly of ribosomes with their export to the cytoplasm. Assembly factors Rpf2 and Rrs1 are necessary for incorporation of 5S rRNA and ribosomal proteins rpL5, rpL10, and rpL11 into 90S preribosomes in the nucleolus. Subsequently, nuclear export receptor Mex67-Mtr2 can bind to 5S rRNA in preribosomes, Nmd3 can bind to rpL10 in pre-rRNPs, and Arx1 can associate with nascent ribosomes, in the nucleoplasm. Mex67-Mtr2, Nmd3, and Arx1 then function to direct preribosomes to and through nuclear pores into the cytoplasm. Nmd3 does so via binding to Xpo1/Crm1.
Fig. 4
A general mechanism for proofreading RNP transitions by DExD/H box ATPases. In the kinetic proofreading scheme, k1 (for S→P, shown in green) represents the rate of a chemical reaction, such as exon ligation, a binding event, such as binding of U2 to the branch site consensus, or potentially a conformation change; k2 (for S→SR, shown in red) represents the rate of rejecting a substrate. Specific discrimination against an incorrect substrate can be established by a slower k1 and/or a faster k2. Note that the DExD/H box ATPase expends energy to reject an incorrect substrate but also expends energy to promote a genuine product (P→P′), if the DExD/H box ATPase functions after, rather than before, the step under inspection. It is currently unclear what determines whether the DExD/H box ATPase acts before or after the proofread step and how the ATPase antagonizes splicing before while promoting splicing after the proofread step.
Similar articles
- The spliceosome: design principles of a dynamic RNP machine.
Wahl MC, Will CL, Lührmann R. Wahl MC, et al. Cell. 2009 Feb 20;136(4):701-18. doi: 10.1016/j.cell.2009.02.009. Cell. 2009. PMID: 19239890 Review. - Ordered and dynamic assembly of single spliceosomes.
Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Hoskins AA, et al. Science. 2011 Mar 11;331(6022):1289-95. doi: 10.1126/science.1198830. Science. 2011. PMID: 21393538 Free PMC article. - Small ribonucleoprotein particle protein SmD3 governs the homeostasis of germline stem cells and the crosstalk between the spliceosome and ribosome signals in Drosophila.
Yu J, Luan X, Yan Y, Qiao C, Liu Y, Zhao D, Xie B, Zheng Q, Wang M, Chen W, Shen C, He Z, Hu X, Huang X, Li H, Chen B, Zheng B, Chen X, Fang J. Yu J, et al. FASEB J. 2019 Jul;33(7):8125-8137. doi: 10.1096/fj.201802536RR. Epub 2019 Mar 28. FASEB J. 2019. PMID: 30921522 - Single-molecule fluorescence-based studies on the dynamics, assembly and catalytic mechanism of the spliceosome.
Warnasooriya C, Rueda D. Warnasooriya C, et al. Biochem Soc Trans. 2014 Aug;42(4):1211-8. doi: 10.1042/BST20140105. Biochem Soc Trans. 2014. PMID: 25110027 Review.
Cited by
- Nucleomorph Genome Sequences of Two Chlorarachniophytes, Amorphochlora amoebiformis and Lotharella vacuolata.
Suzuki S, Shirato S, Hirakawa Y, Ishida K. Suzuki S, et al. Genome Biol Evol. 2015 May 22;7(6):1533-45. doi: 10.1093/gbe/evv096. Genome Biol Evol. 2015. PMID: 26002880 Free PMC article. - Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae.
Talkish J, Zhang J, Jakovljevic J, Horsey EW, Woolford JL Jr. Talkish J, et al. Nucleic Acids Res. 2012 Sep 1;40(17):8646-61. doi: 10.1093/nar/gks609. Epub 2012 Jun 26. Nucleic Acids Res. 2012. PMID: 22735702 Free PMC article. - Global Identification of RNA-Binding Proteins in Bacteria.
Stenum TS, Holmqvist E. Stenum TS, et al. Methods Mol Biol. 2024;2741:347-361. doi: 10.1007/978-1-0716-3565-0_18. Methods Mol Biol. 2024. PMID: 38217662 - Intrinsic disorder in the human spliceosomal proteome.
Korneta I, Bujnicki JM. Korneta I, et al. PLoS Comput Biol. 2012;8(8):e1002641. doi: 10.1371/journal.pcbi.1002641. Epub 2012 Aug 9. PLoS Comput Biol. 2012. PMID: 22912569 Free PMC article. - Optimizing ring assembly reveals the strength of weak interactions.
Deeds EJ, Bachman JA, Fontana W. Deeds EJ, et al. Proc Natl Acad Sci U S A. 2012 Feb 14;109(7):2348-53. doi: 10.1073/pnas.1113095109. Epub 2012 Jan 30. Proc Natl Acad Sci U S A. 2012. PMID: 22308356 Free PMC article.
References
- Decatur W, Fournier MJ. RNA guided nucleotide modification of ribosomal and other RNAs. J Biol Chem. 2003;278:695–698. - PubMed
- Schafer T, Maco B, Petfalski E, Tollervey D, Bottcher B, Aebi M, Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. In vitro assays with purified preribosomes revealed that phosphorylation then dephosphorylation of ribosomal protein rpS3 caused it to be more stably associated with preribosomes. This cycle of modifications of rpS3, required for subunit biogenesis, was also correlated with structural rearrangements of the pre-40S particle, visible by cryo-EM. Mature 40S particles contain a “beak” structure, caused by protruding helix 33 of the 18S rRNA, whereas pre-40S particles lack the structure. The protein kinase Hrr25 was found to be associated with these preribosomes and required in vivo for this phosphorylation of rpS3 and maturation of pre-40S particles. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources