Theoretical limits on errors and acquisition rates in localizing switchable fluorophores - PubMed (original) (raw)
Theoretical limits on errors and acquisition rates in localizing switchable fluorophores
Alexander R Small. Biophys J. 2009 Jan.
Abstract
A variety of recent imaging techniques are able to beat the diffraction limit in fluorescence microcopy by activating and localizing subsets of the fluorescent molecules in the specimen, and repeating this process until all of the molecules have been imaged. In these techniques there is a tradeoff between speed (activating more molecules per imaging cycle) and error rates (activating more molecules risks producing overlapping images that hide information on molecular positions), and so intelligent image processing approaches are needed to identify and reject overlapping images. We introduce here a formalism for defining error rates, derive a general relationship between error rates, image acquisition rates, and the performance characteristics of the image processing algorithms, and show that there is a minimum acquisition time irrespective of algorithm performance. We also consider algorithms that can infer molecular positions from images of overlapping blurs, and derive the dependence of the minimum acquisition time on algorithm performance.
Figures
Figure 1
Concept of a rejection algorithm. Open and solid circles represent activated and dark molecules. When diffraction-blurred images of two molecules don't overlap, or when only one molecule is activated, the image is accepted with probability _f_1. When blurs from two activated molecules overlap, the algorithm rejects the image with probability _f_2.
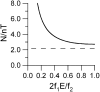
Figure 2
Relationship between number of activation cycles required for a given error rate E and rejection algorithm performance characteristics _f_1 and _f_2.
Similar articles
- Parameter-free image resolution estimation based on decorrelation analysis.
Descloux A, Grußmayer KS, Radenovic A. Descloux A, et al. Nat Methods. 2019 Sep;16(9):918-924. doi: 10.1038/s41592-019-0515-7. Epub 2019 Aug 26. Nat Methods. 2019. PMID: 31451766 - Depth-variant maximum-likelihood restoration for three-dimensional fluorescence microscopy.
Preza C, Conchello JA. Preza C, et al. J Opt Soc Am A Opt Image Sci Vis. 2004 Sep;21(9):1593-601. doi: 10.1364/josaa.21.001593. J Opt Soc Am A Opt Image Sci Vis. 2004. PMID: 15384425 - Analysing errors in single-molecule localisation microscopy.
Costello I, Cox S. Costello I, et al. Int J Biochem Cell Biol. 2021 May;134:105931. doi: 10.1016/j.biocel.2021.105931. Epub 2021 Feb 17. Int J Biochem Cell Biol. 2021. PMID: 33609748 - 3D deconvolution microscopy.
Biggs DS. Biggs DS. Curr Protoc Cytom. 2010 Apr;Chapter 12:Unit 12.19.1-20. doi: 10.1002/0471142956.cy1219s52. Curr Protoc Cytom. 2010. PMID: 20373494 Review. - Microscopy beyond the diffraction limit using actively controlled single molecules.
Moerner WE. Moerner WE. J Microsc. 2012 Jun;246(3):213-20. doi: 10.1111/j.1365-2818.2012.03600.x. Epub 2012 Apr 12. J Microsc. 2012. PMID: 22582796 Free PMC article. Review.
Cited by
- Estimating the dynamic range of quantitative single-molecule localization microscopy.
Nino DF, Milstein JN. Nino DF, et al. Biophys J. 2021 Sep 21;120(18):3901-3910. doi: 10.1016/j.bpj.2021.08.024. Epub 2021 Aug 24. Biophys J. 2021. PMID: 34437847 Free PMC article. - Fluorescence Polarization Control for On-Off Switching of Single Molecules at Cryogenic Temperatures.
Hulleman CN, Huisman M, Moerland RJ, Grünwald D, Stallinga S, Rieger B. Hulleman CN, et al. Small Methods. 2018 Sep 11;2(9):1700323. doi: 10.1002/smtd.201700323. Epub 2018 Apr 30. Small Methods. 2018. PMID: 31240238 Free PMC article. - Resolution limit of image analysis algorithms.
Cohen EAK, Abraham AV, Ramakrishnan S, Ober RJ. Cohen EAK, et al. Nat Commun. 2019 Feb 15;10(1):793. doi: 10.1038/s41467-019-08689-x. Nat Commun. 2019. PMID: 30770826 Free PMC article. - DLBI: deep learning guided Bayesian inference for structure reconstruction of super-resolution fluorescence microscopy.
Li Y, Xu F, Zhang F, Xu P, Zhang M, Fan M, Li L, Gao X, Han R. Li Y, et al. Bioinformatics. 2018 Jul 1;34(13):i284-i294. doi: 10.1093/bioinformatics/bty241. Bioinformatics. 2018. PMID: 29950012 Free PMC article. - Quantum correlation enhanced super-resolution localization microscopy enabled by a fibre bundle camera.
Israel Y, Tenne R, Oron D, Silberberg Y. Israel Y, et al. Nat Commun. 2017 Mar 13;8:14786. doi: 10.1038/ncomms14786. Nat Commun. 2017. PMID: 28287167 Free PMC article.
References
- Abbe E. Contributions to the theory of the microscope and microscopic perception. Archiv. Fur Mikroskopische Anatomic. 1873;9:413–468.
- Hell S.W. Far-field optical nanoscopy. Science. 2007;316:1153–1158. - PubMed
- Lidke K., Rieger B., Jovin T., Heintzmann R. Superresolution by localization of quantum dots using blinking statistics. Opt. Express. 2005;13:7052–7062. - PubMed
- Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources