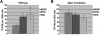ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells - PubMed (original) (raw)
ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells
Monika J Stankiewicz et al. Blood. 2009.
Abstract
ETS2 and ERG are transcription factors, encoded on human chromosome 21 (Hsa21), that have been implicated in human cancer. People with Down syndrome (DS), who are trisomic for Hsa21, are predisposed to acute megakaryoblastic leukemia (AMKL). DS-AMKL blasts harbor a mutation in GATA1, which leads to loss of full-length protein but expression of the GATA-1s isoform. To assess the consequences of ETS protein misexpression on megakaryopoiesis, we expressed ETS2, ERG, and the related protein FLI-1 in wild-type and Gata1 mutant murine fetal liver progenitors. These studies revealed that ETS2, ERG, and FLI-1 facilitated the expansion of megakaryocytes from wild-type, Gata1-knockdown, and Gata1s knockin progenitors, but none of the genes could overcome the differentiation block characteristic of the Gata1-knockdown megakaryocytes. Although overexpression of ETS proteins increased the proportion of CD41(+) cells generated from Gata1s-knockin progenitors, their expression led to a significant reduction in the more mature CD42 fraction. Serial replating assays revealed that overexpression of ERG or FLI-1 immortalized Gata1-knockdown and Gata1s knockin, but not wild-type, fetal liver progenitors. Immortalization was accompanied by activation of the JAK/STAT pathway, commonly seen in megakaryocytic malignancies. These findings provide evidence for synergy between alterations in GATA-1 and overexpression of ETS proteins in aberrant megakaryopoiesis.
Figures
Figure 1
ERG and ETS promote megakaryopoiesis in wild-type fetal liver progenitors. (A) Fetal liver cells transduced with MIGR1, MIGR1-ETS2, or MIGR1-ERG were differentiated for 72 hours and the GFP+ fraction was assessed for CD41 expression by flow cytometry. (B) The degree of polyploidization was determined by DAPI staining of GFP, CD41 double-positive differentiated fetal liver cells. (C) CD42 expression within the GFP-positive population was assessed by flow cytometry. Fold changes for CD41, percentage of cells with DNA content of 8N or more, and CD42 expression in the transduced cells relative to MIGR1 control-infected cells are shown to the right of the representative flow plots. Graphs display average fold changes plus or minus standard error. * P less than .02. For all experiments, n = 4. (D) qRT-PCR analysis of expression of genes that characterize megakaryocyte maturation in wild-type lineage-depleted fetal liver hematopoietic progenitors transduced with MIGR1, MIGR1-ETS2, and MIGR1-ERG. Data depict mean plus or minus standard error for a minimum of 3 independent experiments performed in triplicate. * P less than .05.
Figure 2
ERG and ETS promote megakaryopoiesis in _Gata1_-knockdown progenitors but cannot overcome block in maturation associated with a GATA-1 deficiency. (A) _Gata1_-knockdown fetal liver cells transduced with MIGR1, MIGR1-ETS2, or MIGR1-ERG were differentiated for 72 hours and the GFP+ fraction was assessed for CD41 expression by flow cytometry. (B) The degree of polyploidization was determined by DAPI staining of GFP, CD41 double-positive differentiated fetal liver cells. (C) CD42 expression within the GFP-positive population was assessed by flow cytometry. Fold changes for CD41, percentage of cells with DNA content of 8N or more, and CD42 expression in the transduced cells relative to MIGR1 control-infected cells are shown to the right of the representative flow plots. Graphs display average fold changes plus or minus SE. * P less than .01. For all experiments, n = 4. (D) qRT-PCR analysis of expression of genes that characterize megakaryocyte maturation in _Gata1_-knockdown lineage-depleted fetal liver hematopoietic progenitors transduced with MIGR1, MIGR1-ETS2, and MIGR1-ERG. Data depict mean plus or minus SE for a minimum of 3 independent experiments performed in triplicate. * P less than .05.
Figure 3
ETS2, ERG, and FLI-1 promote megakaryopoiesis in Gata1s knockin progenitors. (A) Gata1s knockin fetal liver cells transduced with MIGR1, MIGR1-ETS2, MIGR1-ERG, or MIGR1-FLI-1 were differentiated for 72 hours and the GFP+ fraction was assessed for CD41 expression by flow cytometry. (B) The degree of polyploidization was determined by DAPI staining of GFP, CD41 double-positive differentiated fetal liver cells. (C) CD42 expression within the GFP-positive population was assessed by flow cytometry. (D) The degree of polyploidization was determined by DAPI staining of GFP, CD42 double-positive differentiated fetal liver cells. Fold changes for CD41 (E), CD42 (F) expression, and percentage of cells with DNA content of 8N or more (G) in the transduced cells relative to MIGR1 control-infected cells are shown below the representative flow plots. Graphs display average fold changes plus or minus SE. * P less than or equal to .03. For all experiments, n = 3.
Figure 4
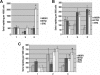
ETS2 and ERG promote CFU-Mk formation from wild-type but not _Gata1_-knockdown progenitors. (A) The number of megakaryocyte colonies generated by GFP+ wild-type fetal liver progenitors transduced with MIGR1, MIGR1-ETS2, or MIGR1-ERG. * P less than .05. (B) The number of megakaryocyte colonies generated by GFP+ _Gata1_-knockdown fetal liver progenitors transduced with MIGR1, MIGR1-ETS2, or MIGR1-ERG. Note that none of the differences are statistically significant.
Figure 5
Overexpression of ETS2 and ERG promote immortalization of _Gata1_-knockdown and Gata1s knockin fetal liver hematopoietic progenitors. (A) The numbers of total colonies derived from plating sorted GFP+ wild-type fetal liver progenitors transduced with MIGR1, MIGR1-ETS2, or MIGR1-ERG for 3 generations. Ten thousand cells were plated each time. No differences were significant except for the ERG third replating. P value less than .03; n = 3 in all experiments. (B) The numbers of total colonies derived from plating sorted GFP+ _Gata1_-knockdown fetal liver progenitors transduced with MIGR1, MIGR1-ETS2, or MIGR1-ERG for 3 generations. Five thousand cells were plated each time. * P less than .05. (C) The numbers of total colonies derived from plating sorted GFP+ Gata1s knockin fetal liver progenitors transduced with MIGR1, MIGR1-ETS2, MIGR1-ERG, or MIGR1-FLI1 for 3 generations. Five thousand cells were plated each time. * P less than or equal to .05.
Figure 6
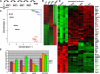
Expression profiling reveals that ERG and FLI-1 have signatures that are distinct from both ETS2 and MIGR1. (A) RNA for genome-wide expression profiling was extracted from fourth-generation _Gata1_-knockdown colonies transduced with the MIGR1, MIGR1-ERG, MIGR1-ETS2, and MIGR1-FLI-1. (B) Heat map generated by unsupervised hierarchic clustering reveals that 726 genes discriminate ETS2-, ERG-, and FLI1-overexpressing colonies from MIGR1-transduced cells. (C) Principal component analysis reveals that the ERG and FLI-1 signatures are close to each other and are well separated from MIGR1 and ETS2. This implies that ERG and FLI-1 are more similar to each other than to MIGR1. (D) Heat map showing the top 50 hematopoiesis genes that are differentially expressed in the samples. Red indicates increased expression by 2-fold or more, whereas green depicts decreased expression by 2-fold or more. Genes within blue boxes are those whose expression differs among the ETS family–transduced cells. (E) qRT-PCR confirmation of several differentially expressed genes in the colonies derived from _Gata1_-knockdown fetal liver hematopoietic cells. Graph depicts mean plus or minus SE for a minimum of 3 independent experiments performed in triplicate. All differences are significant at the 5% level.
Figure 7
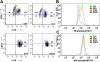
ETS-, ERG-, and FLI-1–immortalized cells express CD41 and c-kit and show enhanced STAT-3 phosphorylation. (A) Comparison of CD41 and c-kit expression in ETS2-, ERG-, and FLI-1–overexpressing _Gata1_-knockdown fetal liver hematopoietic cells isolated from methylcellulose after 4 weeks of culture. Plots are representative of 3 independent experiments. Significance levels for differences for ERG and FLI-1 in comparison with MIGR1 are for CD41 expression (singly positive) only: ERG (P < .01), FLI-1 (P < .04); for CD41/c-kit double-positive gate: ERG (P < .02); c-kit expression: ERG (P < .008), FLI-1 (P < .03). (B,C) Comparison of phospho-STAT3 and phospho-STAT5(a/b) staining of fourth-generation ETS2-, ERG-, and FLI-1-overexpressing _Gata1_-knockdown fetal liver hematopoietic cells. Significance levels for differences in phospho-STAT3 signaling in comparison with MIGR1: ERG (P < .01), FLI-1 (P < .009), and ETS2 (not significant). P values were calculated by comparing the percentage of cells stained for phospho-STAT3 in the different groups for 3 independent experiments.
Comment in
- New GATA1 mutation in codon 2 leads to the earliest known premature stop codon in transient myeloproliferative disorder.
Hoeller S, Bihl MP, Tzankov A, Kuehne T, Potthoff C, Bruder E. Hoeller S, et al. Blood. 2009 Oct 22;114(17):3717-8. doi: 10.1182/blood-2009-07-233833. Blood. 2009. PMID: 19850750 No abstract available.
Similar articles
- ERG is a megakaryocytic oncogene.
Salek-Ardakani S, Smooha G, de Boer J, Sebire NJ, Morrow M, Rainis L, Lee S, Williams O, Izraeli S, Brady HJ. Salek-Ardakani S, et al. Cancer Res. 2009 Jun 1;69(11):4665-73. doi: 10.1158/0008-5472.CAN-09-0075. Cancer Res. 2009. PMID: 19487285 - The role of the proto-oncogene ETS2 in acute megakaryocytic leukemia biology and therapy.
Ge Y, LaFiura KM, Dombkowski AA, Chen Q, Payton SG, Buck SA, Salagrama S, Diakiw AE, Matherly LH, Taub JW. Ge Y, et al. Leukemia. 2008 Mar;22(3):521-9. doi: 10.1038/sj.leu.2405066. Epub 2007 Dec 20. Leukemia. 2008. PMID: 18094719 Free PMC article. - Perturbation of fetal hematopoiesis in a mouse model of Down syndrome's transient myeloproliferative disorder.
Birger Y, Goldberg L, Chlon TM, Goldenson B, Muler I, Schiby G, Jacob-Hirsch J, Rechavi G, Crispino JD, Izraeli S. Birger Y, et al. Blood. 2013 Aug 8;122(6):988-98. doi: 10.1182/blood-2012-10-460998. Epub 2013 May 29. Blood. 2013. PMID: 23719302 Free PMC article. - GATA1 in Normal and Pathologic Megakaryopoiesis and Platelet Development.
Takasaki K, Chou ST. Takasaki K, et al. Adv Exp Med Biol. 2024;1459:261-287. doi: 10.1007/978-3-031-62731-6_12. Adv Exp Med Biol. 2024. PMID: 39017848 Review. - Transcription factor GATA-1 and Down syndrome leukemogenesis.
Muntean AG, Ge Y, Taub JW, Crispino JD. Muntean AG, et al. Leuk Lymphoma. 2006 Jun;47(6):986-97. doi: 10.1080/10428190500485810. Leuk Lymphoma. 2006. PMID: 16840187 Review.
Cited by
- Tumorigenesis in Down's syndrome: big lessons from a small chromosome.
Nižetić D, Groet J. Nižetić D, et al. Nat Rev Cancer. 2012 Oct;12(10):721-32. doi: 10.1038/nrc3355. Epub 2012 Sep 21. Nat Rev Cancer. 2012. PMID: 22996602 - The biology of pediatric acute megakaryoblastic leukemia.
Gruber TA, Downing JR. Gruber TA, et al. Blood. 2015 Aug 20;126(8):943-9. doi: 10.1182/blood-2015-05-567859. Epub 2015 Jul 17. Blood. 2015. PMID: 26186939 Free PMC article. Review. - FUS-ERG induces late-onset azacitidine resistance in acute myeloid leukaemia cells.
Asai-Nishishita A, Kawahara M, Tatsumi G, Iwasa M, Fujishiro A, Nishimura R, Minamiguchi H, Kito K, Murata M, Andoh A. Asai-Nishishita A, et al. Sci Rep. 2023 Sep 2;13(1):14454. doi: 10.1038/s41598-023-41230-1. Sci Rep. 2023. PMID: 37660196 Free PMC article. - Mouse models of diseases of megakaryocyte and platelet homeostasis.
Carmichael CL, Alexander WS. Carmichael CL, et al. Mamm Genome. 2011 Aug;22(7-8):449-65. doi: 10.1007/s00335-011-9336-4. Epub 2011 Jun 11. Mamm Genome. 2011. PMID: 21667128 Review. - Acute Megakaryocytic Leukemia.
McNulty M, Crispino JD. McNulty M, et al. Cold Spring Harb Perspect Med. 2020 Feb 3;10(2):a034884. doi: 10.1101/cshperspect.a034884. Cold Spring Harb Perspect Med. 2020. PMID: 31548219 Free PMC article. Review.
References
- Holmes ML, Bartle N, Eisbacher M, Chong BH. Cloning and analysis of the thrombopoietin-induced megakaryocyte-specific glycoprotein VI promoter and its regulation by GATA-1, Fli-1, and Sp1. J Biol Chem. 2002;277:48333–48341. - PubMed
- Gosiengfiao Y, Horvat R, Thompson A. Transcription factors GATA-1 and Fli-1 regulate human HOXA10 expression in megakaryocytic cells. DNA Cell Biol. 2007;26:577–587. - PubMed
- Rainis L, Toki T, Pimanda JE, et al. The proto-oncogene ERG in megakaryoblastic leukemias. Cancer Res. 2005;65:7596–7602. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases