Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs - PubMed (original) (raw)
. 2009 Feb 19;457(7232):1028-32.
doi: 10.1038/nature07759. Epub 2009 Jan 25.
Collaborators
- PMID: 19169241
- PMCID: PMC2719882
- DOI: 10.1038/nature07759
Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs
Affymetrix ENCODE Transcriptome Project et al. Nature. 2009.
Abstract
The transcriptomes of eukaryotic cells are incredibly complex. Individual non-coding RNAs dwarf the number of protein-coding genes, and include classes that are well understood as well as classes for which the nature, extent and functional roles are obscure. Deep sequencing of small RNAs (<200 nucleotides) from human HeLa and HepG2 cells revealed a remarkable breadth of species. These arose both from within annotated genes and from unannotated intergenic regions. Overall, small RNAs tended to align with CAGE (cap-analysis of gene expression) tags, which mark the 5' ends of capped, long RNA transcripts. Many small RNAs, including the previously described promoter-associated small RNAs, appeared to possess cap structures. Members of an extensive class of both small RNAs and CAGE tags were distributed across internal exons of annotated protein coding and non-coding genes, sometimes crossing exon-exon junctions. Here we show that processing of mature mRNAs through an as yet unknown mechanism may generate complex populations of both long and short RNAs whose apparently capped 5' ends coincide. Supplying synthetic promoter-associated small RNAs corresponding to the c-MYC transcriptional start site reduced MYC messenger RNA abundance. The studies presented here expand the catalogue of cellular small RNAs and demonstrate a biological impact for at least one class of non-canonical small RNAs.
Figures
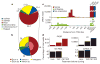
Figure 1. Genomic distribution of small RNAs
a, Annotation of sRNAs from sequencing. ‘Rest’ represents unannotated sRNAs filtered for mitochondria, chromosome Y, repeats and known sRNAs. miRNA, microRNA; ncRNA, non-coding RNA. (Collapsed data are shown.) b, Mapping of unannotated sequences to annotated genomic landmarks. as, antisense to corresponding transcript; s, sense. (Collapsed data.) Shaded inner portions represent the fraction that are PASRs. c, Distribution of sRNAs over TSSs. Orientation is with respect to the long transcript. Antisense sRNAs are plotted with a different _y_-axis beneath. (Uncollapsed data.) d, Characterization of PASR 5′ ends. Untr., untreated. Sequences corresponding to the 5′ end of U4, 5S rRNA and mir-21 were extracted as controls. (Uncollapsed data.)
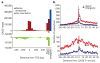
Figure 2. Correlation of sRNAs and CAGE tags
a, Distribution of CAGE tags over annotated TSSs. Orientation is with respect to the long transcript. Antisense sRNAs are plotted with a different _y_-axis beneath. (Uncollapsed data.) b, Distribution of PASR (top) and non-PASR sRNAs (bottom) around CAGE tag 5′ ends. The distance to closest short RNA5′ end was plotted for each CAGE tag. (Collapsed data.)
Figure 3. Correlation between CAGE tags, sRNAs and internal exons of annotated transcripts
a, Left: distribution of mapped CAGE tag 5′ ends across internal exons. Exon length was normalized to 100 segments. Right: distribution of CAGE tags not mapping to the genome but mapping to exon–exon junctions (EEJ) of internal exons. b, Prevalence of internal CAGE tags. Black line represents the maximum expected exons in random samplings (see Methods). Colour corresponds to number of transcripts represented by each data point. c, CAGE tag and sRNA coverage of the APOB gene. sRNAs from cap-immunoprecipitation (IP) are shown separately. Histone H3 acetylation (H3AC) pattern is shown below. Two internal exons are magnified. d, Characterization of libraries from anti-cap-immunoprecipitated RNA. Top panel: representation of sRNAs in total and IP libraries (uncollapsed data). For all but the U4 fraction, uniquely mapping sequences were considered. Bottom panel: distance to closest sRNA 5′ end from CAGE 5′ end in internal exons (collapsed data).
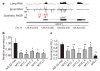
Figure 4. Regulation of gene expression by PASRs
a, Expression profile of the MYC locus. The long and short RNA profile of HeLa cells on Affymetrix tiling arrays. Red rectangles indicate the designed synthetic PASRs (MYC_1–5 are denoted by numbers and sequence information is provided in Supplementary Table 2) corresponding to peaks in the sRNA array profile. b, MYC mRNA expression levels in HeLa cells as measured by quantitative PCR with reverse transcription (n = 3, P values <0.01). c, Effects of PASR transfections on a MYC-responsive luciferase transcriptional reporter in HeLa cells was measured as relative light units (RLU) (n = 2, *P <0.01, **P <0.001). For reference, a control 33-mer and an siRNA directed against luciferase (siGL3) are shown.
Figure 5. A proposed model for the metabolism of genic transcripts into a diversity of long and short RNAs
Transcription of a genic region results in a precursor long RNA containing a 5′ cap structure, as shown by asterisks. After processing into spliced RNAs, protein-coding RNAs are destined either to be translated or to be further processed. This further processing entails cleavage followed in some cases by addition of a 5′ modification, possibly a cap structure. Additional cleavage of these intermediate products can generate a class of short RNAs, some also bearing a cap structure. lRNAs, long RNAs.
Comment in
- Molecular biology: The long and short of RNAs.
Carninci P. Carninci P. Nature. 2009 Feb 19;457(7232):974-5. doi: 10.1038/457974b. Nature. 2009. PMID: 19225515 No abstract available.
Similar articles
- Molecular biology: The long and short of RNAs.
Carninci P. Carninci P. Nature. 2009 Feb 19;457(7232):974-5. doi: 10.1038/457974b. Nature. 2009. PMID: 19225515 No abstract available. - Expression of mitochondrial protein-coding genes in Tetrahymena pyriformis.
Edqvist J, Burger G, Gray MW. Edqvist J, et al. J Mol Biol. 2000 Mar 24;297(2):381-93. doi: 10.1006/jmbi.2000.3530. J Mol Biol. 2000. PMID: 10715208 - Relating underrepresented genomic DNA patterns and tiRNAs: the rule behind the observation and beyond.
Cserzo M, Turu G, Varnai P, Hunyady L. Cserzo M, et al. Biol Direct. 2010 Sep 22;5:56. doi: 10.1186/1745-6150-5-56. Biol Direct. 2010. PMID: 20860791 Free PMC article. - Maturation of 5' ends of plant mitochondrial RNAs.
Binder S, Stoll K, Stoll B. Binder S, et al. Physiol Plant. 2016 Jul;157(3):280-8. doi: 10.1111/ppl.12423. Epub 2016 Mar 23. Physiol Plant. 2016. PMID: 26833432 Review. - Exploring long non-coding RNAs through sequencing.
Atkinson SR, Marguerat S, Bähler J. Atkinson SR, et al. Semin Cell Dev Biol. 2012 Apr;23(2):200-5. doi: 10.1016/j.semcdb.2011.12.003. Epub 2011 Dec 20. Semin Cell Dev Biol. 2012. PMID: 22202731 Review.
Cited by
- Transcription start site evolution in Drosophila.
Main BJ, Smith AD, Jang H, Nuzhdin SV. Main BJ, et al. Mol Biol Evol. 2013 Aug;30(8):1966-74. doi: 10.1093/molbev/mst085. Epub 2013 May 6. Mol Biol Evol. 2013. PMID: 23649539 Free PMC article. - A meta-analysis of the genomic and transcriptomic composition of complex life.
Liu G, Mattick JS, Taft RJ. Liu G, et al. Cell Cycle. 2013 Jul 1;12(13):2061-72. doi: 10.4161/cc.25134. Epub 2013 Jun 6. Cell Cycle. 2013. PMID: 23759593 Free PMC article. - Structure and function of long noncoding RNAs in epigenetic regulation.
Mercer TR, Mattick JS. Mercer TR, et al. Nat Struct Mol Biol. 2013 Mar;20(3):300-7. doi: 10.1038/nsmb.2480. Nat Struct Mol Biol. 2013. PMID: 23463315 Review. - Integration of complex data sources to provide biologic insight into pulmonary vascular disease (2015 Grover Conference Series).
Brittain EL, Chan SY. Brittain EL, et al. Pulm Circ. 2016 Sep;6(3):251-60. doi: 10.1086/686995. Pulm Circ. 2016. PMID: 27683602 Free PMC article. Review. - Dynamics of microRNA expression during mouse prenatal development.
Rahmanian S, Murad R, Breschi A, Zeng W, Mackiewicz M, Williams B, Davis CA, Roberts B, Meadows S, Moore D, Trout D, Zaleski C, Dobin A, Sei LH, Drenkow J, Scavelli A, Gingeras TR, Wold BJ, Myers RM, Guigó R, Mortazavi A. Rahmanian S, et al. Genome Res. 2019 Nov;29(11):1900-1909. doi: 10.1101/gr.248997.119. Epub 2019 Oct 23. Genome Res. 2019. PMID: 31645363 Free PMC article.
References
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nature Rev Genet. 2007;8:413–423. - PubMed
- Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. - PubMed
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous