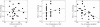Prospects for de novo phasing with de novo protein models - PubMed (original) (raw)
Prospects for de novo phasing with de novo protein models
Rhiju Das et al. Acta Crystallogr D Biol Crystallogr. 2009 Feb.
Abstract
The prospect of phasing diffraction data sets ;de novo' for proteins with previously unseen folds is appealing but largely untested. In a first systematic exploration of phasing with Rosetta de novo models, it is shown that all-atom refinement of coarse-grained models significantly improves both the model quality and performance in molecular replacement with the Phaser software. 15 new cases of diffraction data sets that are unambiguously phased with de novo models are presented. These diffraction data sets represent nine space groups and span a large range of solvent contents (33-79%) and asymmetric unit copy numbers (1-4). No correlation is observed between the ease of phasing and the solvent content or asymmetric unit copy number. Instead, a weak correlation is found with the length of the modeled protein: larger proteins required somewhat less accurate models to give successful molecular replacement. Overall, the results of this survey suggest that de novo models can phase diffraction data for approximately one sixth of proteins with sizes of 100 residues or less. However, for many of these cases, ;de novo phasing with de novo models' requires significant investment of computational power, much greater than 10(3) CPU days per target. Improvements in conformational search methods will be necessary if molecular replacement with de novo models is to become a practical tool for targets without homology to previously solved protein structures.
Figures
Figure 1
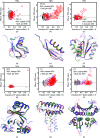
New examples of successful molecular replacement with Rosetta de novo models. (a)–(c) and (g)–(i) display correlations of Phaser translation-function Z score (TFZ) with model accuracy (the fraction of Cα atoms within 1 Å of the crystal structure). For each target, the displayed subsets are 200 randomly selected all-atom refined models (black) and 200 models with lowest energy from the 100 CPU-day low-resolution set (gray), from the 100 CPU-day all-atom refined set (magenta) and from the large-scale all-atom refined set (red). The solid line and dashed line display the mean TFZ scores and a cutoff value five standard deviations above the mean TFZ, respectively, in the randomly chosen models. Larger open circles indicate Phaser solutions with correct orientations in the unit cell (see text). (d)–(f) and (j)–(l) give overlays corresponding to each plot in (a)–(c) and (g)–(i), respectively, of the least accurate model that passes the TFZ cutoff value (red, partly transparent), nearly complete models built by ARP/wARP after molecular replacement (green) and the crystal structure (blue). In some cases, the modeled sequence did not include terminal segments present in the crystal structures [see red structures in (d)–(f) and (j)–(l)].
Figure 2
Dependence of de novo phasing on crystallographic parameters. The ease of phasing is estimated as the minimal accuracy required for successful molecular replacement (minimum F 1 Å, the fraction of Cα atoms within 1 Å of the crystal structure). No correlation is observed with the crystal solvent content (a) or the number of molecules in each asymmetric unit (b), but a statistically significant correlation is found with the number of residues in the molecular-replacement model (c). See also Table 2 ▶.
Similar articles
- Accelerating ab initio phasing with de novo models.
Shrestha R, Berenger F, Zhang KY. Shrestha R, et al. Acta Crystallogr D Biol Crystallogr. 2011 Sep;67(Pt 9):804-12. doi: 10.1107/S090744491102779X. Epub 2011 Aug 9. Acta Crystallogr D Biol Crystallogr. 2011. PMID: 21904033 - Error-estimation-guided rebuilding of de novo models increases the success rate of ab initio phasing.
Shrestha R, Simoncini D, Zhang KY. Shrestha R, et al. Acta Crystallogr D Biol Crystallogr. 2012 Nov;68(Pt 11):1522-34. doi: 10.1107/S0907444912037961. Epub 2012 Oct 18. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 23090401 - A fragmentation and reassembly method for ab initio phasing.
Shrestha R, Zhang KY. Shrestha R, et al. Acta Crystallogr D Biol Crystallogr. 2015 Feb;71(Pt 2):304-12. doi: 10.1107/S1399004714025449. Epub 2015 Jan 23. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25664740 - Implications of AlphaFold2 for crystallographic phasing by molecular replacement.
McCoy AJ, Sammito MD, Read RJ. McCoy AJ, et al. Acta Crystallogr D Struct Biol. 2022 Jan 1;78(Pt 1):1-13. doi: 10.1107/S2059798321012122. Epub 2022 Jan 1. Acta Crystallogr D Struct Biol. 2022. PMID: 34981757 Free PMC article. Review. - Acknowledging Errors: Advanced Molecular Replacement with Phaser.
McCoy AJ. McCoy AJ. Methods Mol Biol. 2017;1607:421-453. doi: 10.1007/978-1-4939-7000-1_18. Methods Mol Biol. 2017. PMID: 28573584 Review.
Cited by
- Computed structures of point deletion mutants and their enzymatic activities.
Berrondo M, Gray JJ. Berrondo M, et al. Proteins. 2011 Oct;79(10):2844-60. doi: 10.1002/prot.23109. Epub 2011 Aug 23. Proteins. 2011. PMID: 21905110 Free PMC article. - Assessment of protein structure refinement in CASP9.
MacCallum JL, Pérez A, Schnieders MJ, Hua L, Jacobson MP, Dill KA. MacCallum JL, et al. Proteins. 2011;79 Suppl 10(Suppl 10):74-90. doi: 10.1002/prot.23131. Epub 2011 Aug 30. Proteins. 2011. PMID: 22069034 Free PMC article. - Residue contacts predicted by evolutionary covariance extend the application of ab initio molecular replacement to larger and more challenging protein folds.
Simkovic F, Thomas JM, Keegan RM, Winn MD, Mayans O, Rigden DJ. Simkovic F, et al. IUCrJ. 2016 Jun 15;3(Pt 4):259-70. doi: 10.1107/S2052252516008113. eCollection 2016 Jul 1. IUCrJ. 2016. PMID: 27437113 Free PMC article. - Practically useful: what the Rosetta protein modeling suite can do for you.
Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J. Kaufmann KW, et al. Biochemistry. 2010 Apr 13;49(14):2987-98. doi: 10.1021/bi902153g. Biochemistry. 2010. PMID: 20235548 Free PMC article. Review. - Modeling large regions in proteins: applications to loops, termini, and folding.
Adhikari AN, Peng J, Wilde M, Xu J, Freed KF, Sosnick TR. Adhikari AN, et al. Protein Sci. 2012 Jan;21(1):107-21. doi: 10.1002/pro.767. Epub 2011 Dec 5. Protein Sci. 2012. PMID: 22095743 Free PMC article.
References
- Blow, D. M. & Rossmann, M. G. (1961). Acta Cryst.14, 1195–1202.
- Blum, B., Jordan, M. I., Kim, D., Das, R., Bradley, P. & Baker, D. (2007). In Advances in Neural Information Processing Systems, Vol. 20, edited by J. Platt, D. Koller, Y. Singer & A. McCallum. Cambridge, USA: MIT Press.
- Bradley, P., Misura, K. M. & Baker, D. (2005). Science, 309, 1868–1871. - PubMed
- Chen, Y. W., Dodson, E. J. & Kleywegt, G. J. (2000). Structure, 8, R213–R220. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources