Essential and overlapping functions for mammalian Argonautes in microRNA silencing - PubMed (original) (raw)
Essential and overlapping functions for mammalian Argonautes in microRNA silencing
Hong Su et al. Genes Dev. 2009.
Abstract
MicroRNA (miRNA) silencing fine-tunes protein output and regulates diverse biological processes. Argonaute (Ago) proteins are the core effectors of the miRNA pathway. In lower organisms, multiple Agos have evolved specialized functions for distinct RNA silencing pathways. However, the roles of mammalian Agos have not been well characterized. Here we show that mouse embryonic stem (ES) cells deficient for Ago1-4 are completely defective in miRNA silencing and undergo apoptosis. In miRNA silencing-defective ES cells, the proapoptotic protein Bim, a miRNA target, is increased, and up-regulation of Bim is sufficient to induce ES cell apoptosis. Expression of activated Akt inhibits Bim expression and partially rescues the growth defect in Ago-deficient ES cells. Furthermore, reintroduction of any single Ago into Ago-deficient cells is able to rescue the endogenous miRNA silencing defect and apoptosis. Consistent with this, each Ago is functionally equivalent with bulged miRNA duplexes for translational repression, whereas Ago1 and Ago2 appear to be more effective at utilizing perfectly matched siRNAs. Thus, our results demonstrate that mammalian Agos all contribute to miRNA silencing, and individual Agos have largely overlapping functions in this process.
Figures
Figure 1.
Generation of inducible knockout ES cells for all four Ago proteins. (A) A genetic strategy to generate inducible ES cells deficient for Ago1–4. Multiple steps of gene targeting and Cre-mediated excision were employed to first generate B9 ES cells (Ago1, Ago3, _Ago4_−/−). In the presence of floxed hAgo2 transgene, both mAgo2 alleles were deleted by targeting in E9 ES cells. A CreERT2 transgene was finally introduced to generate the inducible E7 line. (B) RT–PCR analyses confirm that the endogenous Ago transcripts are absent in mutant ES cell lines. Note that related PIWI transcripts remain unchanged. (C) Schematic of the 4OH-T-induced excision of the hAgo2 transgene from E7 cells. (D) Western blot analysis showing that hAgo2 is expressed at a lower level in E7 cells compared with the endogenous Ago2 in control ES cells and hAgo2 is absent after 4OH-T induction. (E) RT–PCR analysis on E7 cells showing a decrease in hAgo2 after 2–4 d of 4OH-T treatment. The residual hAgo2 expression reflects an incomplete Cre excision. Based on calculating the number of drug (Bsd)-resistant colonies, we estimated that >80% of E7 cells excised hAgo2 by CreERT2 after 48-h 4OHT treatment and the excision efficiency increases with longer treatments. (UT) Untreated.
Figure 2.
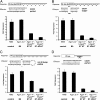
Ago1–4 are required for miRNA-mediated translational repression. (A–C) miRNA-mediated translation repression is defective in _Ago1-4_−/−; hAgo2Δ ES cells. Shown are luciferase assays using two different reporters and three different effectors indicated in A–C. (D) siRNA-mediated mRNA cleavage is defective in _Ago1-4_−/−; hAgo2Δ E7 ES cells. Shown are assays using a firefly luciferase reporter with a single perfect complementary antisense CXCR-binding site in the 3′UTR of Ff-luc using CXCR dsRNA as an effector. All results are shown as means ± SEM from six independent transfections.
Figure 3.
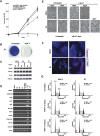
Ago1–4 are essential for the survival of mouse ES cells. (A) Growth curves showing a severe growth defect in _Ago1-4_−/−; hAgo2Δ E7 ES cells. (B) E7 ES cells are not viable after 4OH-T-induced Ago depletion. Shown are methylene blue stains of both untreated and 4OH-T-treated E7 ES cells after five consecutive passages at a ratio of 1:4. (C) The pluripotent transcription factors Oct3/4, Sox2, and Nanog are not down-regulated in Ago mutant ES cell lines E9 and E7 as compared with wild-type (AB2.2). β-tubulin serves as a loading control. (D) RT–PCR analyses on various cell lineage and differentiation markers demonstrate that no abnormal differentiation occurs in Ago1–4 mutant cells. The control is a mixture of RNAs from differentiated embryoid bodies and E12.5 mouse embryos. (E) Phase-contrast images of wild-type and E7 ES cells treated with 4OH-T at various time points. E7 cells begin to die and detach (seen as rounded up bright cells) after 4OH-T treatment. (F) Active caspase-3 staining on wild-type and E7 cells untreated and treated with 4OH-T shows a significant increase of apoptosis in E7-treated cells. (G) FACScan analysis of the cell cycle profiles of wild-type and E7-untreated and -treated ES cells. Note an increase of dying cells (arrow) and a decrease of S-phase cells in 4-d 4OH-T-treated E7 cells.
Figure 4.
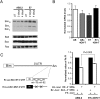
Bim is an endogenous miRNA target in mouse ES cells. (A) Bim protein is up-regulated in Ago-depleted ES cells. Western blots show that three Bim isoforms are increased in miRNA-defective E7 cells. Note other proapoptotic proteins such as Bad, Bax, and miRNA let-7 target Ras remain unchanged in Ago mutant ES cells. (B) RT–quantitative PCR for Bim show no significant change in mRNA levels in Ago-depleted ES cells. (C) The 3′UTR of Bim contains functional binding sites for mir-19 and mir-92, two miRNAs expressed in ES cells. miRNA-mediated repression of a Rr-luc reporter containing a segment of the 3′UTR of Bim was compared with that of a Rr-luc reporter containing a mutant 3′UTR from Bim in both control and Ago-depleted E7 cells. E7 cells were unable to repress the Rr-luc reporter containing the wild-type Bim 3′UTR to the same level as wild-type control (P < 0.003). The data shown are from 12 independent transfections.
Figure 5.
Regulation of ES cell survival by Bim. (A) Schematic of an inducible Bim ES cell line. Upon 4OH-T treatment, CreERT2 excises Neo selection marker and activates Bim transcription. (B) Western blots show that BimEL and BimL proteins are rapidly induced after 4OH-T treatment in two independent inducible ES cell lines. (C) Methylene blue stains show that most Bim-expressing cells are lost 2 d after 4OH-T induction. (D) Bim overexpression induces apoptosis in ES cells. Phase-contrast images and active caspase-3 stains are shown. (E) An activated Akt (myr-Akt) partially rescues cell growth defects of _Ago1-4_−/−; hAgo2Δ E7 cells after five passages with 4OH-T. Western blotting shows that HA-tagged myr-Akt is expressed in rescued E7 cells. Ago2-expressing E7 cells serve as the positive control for a complete rescue. (F) myr-Akt down-regulates Bim transcription in 4OH-T-treated E7 ES cells. Bim mRNA and protein levels are decreased in myr-Akt-rescued E7 cells. Shown are quantitative PCR and Western blot results. (G) The luciferase assay confirms loss of Ago2 and inability to use shRNA to silence a Ff-Luc reporter in myr-Akt-rescued cells. RT–PCR analysis shows that the majority of rescued E7 cells no longer express the floxed hAgo2 as a pool. The residual PCR signal from the pool might reflect a small population of rescued cells that still express hAgo2.
Figure 6.
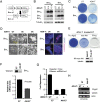
Genetic rescue reveals overlapping functions of individual Ago proteins. (A) Individual Agos rescue 4OH-T-treated E7 cells from apoptosis, whereas controls Ff-luc and Miwi are unable to rescue. Shown are methylene blue stains of indicated ES cells after five passages with or without 4OH-T. (B) Individual HA-tagged Ago1–4 are expressed at a similar level in rescued cell lines with β-tubulin included as a loading control. (C) A cleavage assay shows that the floxed hAgo2 is deleted and confirms that only a single Ago is expressed in each rescued cell line. The shRNA-luc1-mediated mRNA cleavage is measured by dual luciferase assay using Rr-Luc as an internal control. Compared with untreated E7 cells, Ago1-, Ago3-, and Ago4-rescued E7 cells have no cleavage activity, demonstrating that the floxed hAgo2 allele is completely deleted in these cells. Shown are means ± SEM from six independent transfections. (D) Individual Ago proteins are capable of rescuing the endogenous miRNA silencing defect in E7 cells. For the endogenous miRNA target protein Bim, Western blots show that three Bim isoforms are increased in miRNA-defective cells and return to a basal level in individual Ago-rescued E7 cells. Tubulin serves as a loading control.
Figure 7.
Individual Ago proteins have different preferences for miRNAs and siRNAs. (A,B) Different combinations of effectors and luciferase reporters were used in the translational repression assay to compare the loading preference of individual Ago proteins for small RNA duplexes with distinct secondary structures. Shown above each graph are the reporter used and the secondary structure of dsRNA effector. Ago2 and Ago1 are more effective than Ago3 and Ago4 in utilizing siRNA duplexes, while all Agos can repress with miRNA effectors. The experiments in A and B used mir-CXCR and mir-30 reporters, respectively. Shown are means ± SEM from six independent transfections. (C) A summary illustrates the partition of miRNA and siRNA duplexes among different mammalian Ago proteins.
Similar articles
- Generation of an inducible mouse ES cell lines deficient for Argonaute proteins.
Su H, Wang X. Su H, et al. Methods Mol Biol. 2011;725:295-313. doi: 10.1007/978-1-61779-046-1_19. Methods Mol Biol. 2011. PMID: 21528461 - Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway.
Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Ghildiyal M, et al. RNA. 2010 Jan;16(1):43-56. doi: 10.1261/rna.1972910. Epub 2009 Nov 16. RNA. 2010. PMID: 19917635 Free PMC article. - Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54.
Chu CY, Rana TM. Chu CY, et al. PLoS Biol. 2006 Jul;4(7):e210. doi: 10.1371/journal.pbio.0040210. PLoS Biol. 2006. PMID: 16756390 Free PMC article. - From the Argonauts Mythological Sailors to the Argonautes RNA-Silencing Navigators: Their Emerging Roles in Human-Cell Pathologies.
Pantazopoulou VI, Georgiou S, Kakoulidis P, Giannakopoulou SN, Tseleni S, Stravopodis DJ, Anastasiadou E. Pantazopoulou VI, et al. Int J Mol Sci. 2020 Jun 3;21(11):4007. doi: 10.3390/ijms21114007. Int J Mol Sci. 2020. PMID: 32503341 Free PMC article. Review. - The paradigm of miRNA and siRNA influence in Oral-biome.
Sinha A, Bhattacharjee R, Bhattacharya B, Nandi A, Shekhar R, Jana A, Saha K, Kumar L, Patro S, Panda PK, Kaushik NK, Suar M, Verma SK. Sinha A, et al. Biomed Pharmacother. 2023 Mar;159:114269. doi: 10.1016/j.biopha.2023.114269. Epub 2023 Jan 20. Biomed Pharmacother. 2023. PMID: 36682246 Review.
Cited by
- Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter.
Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Chu Y, et al. Nucleic Acids Res. 2010 Nov;38(21):7736-48. doi: 10.1093/nar/gkq648. Epub 2010 Jul 30. Nucleic Acids Res. 2010. PMID: 20675357 Free PMC article. - CD95/Fas ligand induced toxicity.
Haluck-Kangas A, Peter ME. Haluck-Kangas A, et al. Biochem Soc Trans. 2023 Feb 27;51(1):21-29. doi: 10.1042/BST20211187. Biochem Soc Trans. 2023. PMID: 36629505 Free PMC article. - The microRNA processing subunit DGCR8 is required for a T cell-dependent germinal center response.
Daum P, Ottmann SR, Meinzinger J, Schulz SR, Côrte-Real J, Hauke M, Roth E, Schuh W, Mielenz D, Jäck HM, Pracht K. Daum P, et al. Front Immunol. 2022 Dec 16;13:991347. doi: 10.3389/fimmu.2022.991347. eCollection 2022. Front Immunol. 2022. PMID: 36591274 Free PMC article. - Chromatin remodeling by the small RNA machinery in mammalian cells.
Li LC. Li LC. Epigenetics. 2014 Jan;9(1):45-52. doi: 10.4161/epi.26830. Epub 2013 Oct 22. Epigenetics. 2014. PMID: 24149777 Free PMC article. Review. - RNAi-mediated inhibition of HIV-1 by targeting partially complementary viral sequences.
Liu YP, Gruber J, Haasnoot J, Konstantinova P, Berkhout B. Liu YP, et al. Nucleic Acids Res. 2009 Oct;37(18):6194-204. doi: 10.1093/nar/gkp644. Epub 2009 Aug 5. Nucleic Acids Res. 2009. PMID: 19656954 Free PMC article.
References
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. - PubMed
- Bushati N., Cohen S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases