miRNAS in normal and diseased skeletal muscle - PubMed (original) (raw)
Review
miRNAS in normal and diseased skeletal muscle
Iris Eisenberg et al. J Cell Mol Med. 2009 Jan.
Abstract
The last 20 years have witnessed major advances in the understanding of muscle diseases and significant inroads are being made to treat muscular dystrophy. However, no curative therapy is currently available for any of the muscular dystrophies, despite the immense progress made using several approaches and only palliative and symptomatic treatment is available for patients. The discovery of miRNAs as new and important regulators of gene expression is expected to broaden our biological understanding of the regulatory mechanism in muscle by adding another dimension of regulation to the diversity and complexity of gene-regulatory networks. As important regulators of muscle development, unravelling the regulatory circuits involved may be challenging, given that a single miRNA can regulate the expression of many mRNA targets. Although the identification of the regulatory targets of miRNAs in muscle is a challenge, it will be critical for placing them in genetic pathways and biological contexts. Therefore, combining informatics, biochemical and genetic approaches will not only expected to reveal the elucidation of the miRNA regulatory network in skeletal muscle and to bring a better knowledge on muscle tissue regulation but will also raise new opportunities for therapeutic intervention in muscular dystrophies by identifying candidate miRNAs as potential targets for clinical application.
Figures
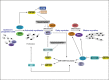
1
miRNA in muscle development. miR-1 and miR-133 along with miR-206 and miR-181 function at the centre of a network of transcription factors to regulate skeletal myoblast proliferation and differentiation. Myocyte enhancer factor-2 (MEF2) and myogenic basic helix loop helix (bHLH) proteins, including myogenic transcription factor MYOD1, regulate their own expression, as well as the expression of downstream muscle structural genes. Additionally, these transcription factors use upstream and intragenic enhancers to activate transcription of bicistronic miR-1/133 clusters encoding miR-1 and miR-133 in differentiated skeletal muscle. miR-1 represses expression of HDAC4 (histone deacetylase 4), a signal-dependent repressor of MEF2 activity, thereby establishing a negative feedback loop to modulate miR-1 and miR-133 expression and promoting myoblast differentiation. As myogenesis progresses from the myoblast stage to the myotube stage, the level of the muscle-specific miR-133 increases and miR-133 represses expression of serum response factor (SRF), a positive regulator of miR-1/133 expression and repressor of myoblast proliferation. Upon differentiation, miR-181 is also up-regulated, resulting in down-regulation of Hox-A11 and in the release of MYOD1 expression. As a result, myogenin (MYOG) and muscle marker proteins including MHC (myosin heavy chain) are up-regulated. In parallel, activation of MYOD1 increases the expression of the primary miR-206 transcript which in turn lead to down-regulation of Follistatin-like 1 (FSTL1) and to the repression of Utrophin (Utrn) expression and through a mechanism that is not yet known promotes muscle differentiation.
Similar articles
- Emerging role of MyomiRs as biomarkers and therapeutic targets in skeletal muscle diseases.
Srivastava S, Rathor R, Singh SN, Suryakumar G. Srivastava S, et al. Am J Physiol Cell Physiol. 2021 Nov 1;321(5):C859-C875. doi: 10.1152/ajpcell.00057.2021. Epub 2021 Sep 29. Am J Physiol Cell Physiol. 2021. PMID: 34586896 Review. - MicroRNAs in the regeneration of skeletal muscle.
Yu X, Zuo Q. Yu X, et al. Front Biosci (Landmark Ed). 2013 Jan 1;18(2):608-15. doi: 10.2741/4124. Front Biosci (Landmark Ed). 2013. PMID: 23276946 Review. - microRNAs and muscle disorders.
Chen JF, Callis TE, Wang DZ. Chen JF, et al. J Cell Sci. 2009 Jan 1;122(Pt 1):13-20. doi: 10.1242/jcs.041723. J Cell Sci. 2009. PMID: 19092056 Free PMC article. Review. - microRNAs: novel components in a muscle gene regulatory network.
Wang H, Sun H, Guttridge DC. Wang H, et al. Cell Cycle. 2009 Jun 15;8(12):1833-7. doi: 10.4161/cc.8.12.8851. Epub 2009 Jun 27. Cell Cycle. 2009. PMID: 19448406 - microRNAs in skeletal muscle differentiation and disease.
Goljanek-Whysall K, Sweetman D, Münsterberg AE. Goljanek-Whysall K, et al. Clin Sci (Lond). 2012 Dec;123(11):611-25. doi: 10.1042/CS20110634. Clin Sci (Lond). 2012. PMID: 22888971 Review.
Cited by
- MiR-351 transiently increases during muscle regeneration and promotes progenitor cell proliferation and survival upon differentiation.
Chen Y, Melton DW, Gelfond JA, McManus LM, Shireman PK. Chen Y, et al. Physiol Genomics. 2012 Nov 1;44(21):1042-51. doi: 10.1152/physiolgenomics.00052.2012. Epub 2012 Sep 11. Physiol Genomics. 2012. PMID: 22968638 Free PMC article. - S100B protein in skeletal muscle regeneration: regulation of myoblast and macrophage functions.
Riuzzi F, Beccafico S, Sorci G, Donato R. Riuzzi F, et al. Eur J Transl Myol. 2016 Feb 23;26(1):5830. doi: 10.4081/ejtm.2016.5830. eCollection 2016 Feb 23. Eur J Transl Myol. 2016. PMID: 27054019 Free PMC article. No abstract available. - Functional and Morphological Improvement of Dystrophic Muscle by Interleukin 6 Receptor Blockade.
Pelosi L, Berardinelli MG, De Pasquale L, Nicoletti C, D'Amico A, Carvello F, Moneta GM, Catizone A, Bertini E, De Benedetti F, Musarò A. Pelosi L, et al. EBioMedicine. 2015 Feb 26;2(4):285-93. doi: 10.1016/j.ebiom.2015.02.014. eCollection 2015 Apr. EBioMedicine. 2015. PMID: 26137572 Free PMC article. - Hmgb3 is regulated by microRNA-206 during muscle regeneration.
Maciotta S, Meregalli M, Cassinelli L, Parolini D, Farini A, Fraro GD, Gandolfi F, Forcato M, Ferrari S, Gabellini D, Bicciato S, Cossu G, Torrente Y. Maciotta S, et al. PLoS One. 2012;7(8):e43464. doi: 10.1371/journal.pone.0043464. Epub 2012 Aug 17. PLoS One. 2012. PMID: 22912879 Free PMC article. - Correlated mRNAs and miRNAs from co-expression and regulatory networks affect porcine muscle and finally meat properties.
Ponsuksili S, Du Y, Hadlich F, Siengdee P, Murani E, Schwerin M, Wimmers K. Ponsuksili S, et al. BMC Genomics. 2013 Aug 5;14:533. doi: 10.1186/1471-2164-14-533. BMC Genomics. 2013. PMID: 23915301 Free PMC article.
References
- Dalkilic I, Kunkel LM. Muscular dystrophies: genes to pathogenesis. Curr Opin Genet Dev. 2003;13:231–8. - PubMed
- Davies KE, Nowak KJ. Molecular mechanisms of muscular dystrophies: old and new players. Nat Rev Mol Cell Biol. 2006;7:762–73. - PubMed
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. - PubMed
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. - PubMed
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the Celegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials