Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis - PubMed (original) (raw)
Comparative Study
doi: 10.1186/1471-2164-10-54.
Matthew T G Holden, James A Leigh, Nicola Lennard, Alexandra Bignell, Andy Barron, Louise Clark, Michael A Quail, John Woodward, Bart G Barrell, Sharon A Egan, Terence R Field, Duncan Maskell, Michael Kehoe, Christopher G Dowson, Neil Chanter, Adrian M Whatmore, Stephen D Bentley, Julian Parkhill
Affiliations
- PMID: 19175920
- PMCID: PMC2657157
- DOI: 10.1186/1471-2164-10-54
Comparative Study
Evidence for niche adaptation in the genome of the bovine pathogen Streptococcus uberis
Philip N Ward et al. BMC Genomics. 2009.
Abstract
Background: Streptococcus uberis, a Gram positive bacterial pathogen responsible for a significant proportion of bovine mastitis in commercial dairy herds, colonises multiple body sites of the cow including the gut, genital tract and mammary gland. Comparative analysis of the complete genome sequence of S. uberis strain 0140J was undertaken to help elucidate the biology of this effective bovine pathogen.
Results: The genome revealed 1,825 predicted coding sequences (CDSs) of which 62 were identified as pseudogenes or gene fragments. Comparisons with related pyogenic streptococci identified a conserved core (40%) of orthologous CDSs. Intriguingly, S. uberis 0140J displayed a lower number of mobile genetic elements when compared with other pyogenic streptococci, however bacteriophage-derived islands and a putative genomic island were identified. Comparative genomics analysis revealed most similarity to the genomes of Streptococcus agalactiae and Streptococcus equi subsp. zooepidemicus. In contrast, streptococcal orthologs were not identified for 11% of the CDSs, indicating either unique retention of ancestral sequence, or acquisition of sequence from alternative sources. Functions including transport, catabolism, regulation and CDSs encoding cell envelope proteins were over-represented in this unique gene set; a limited array of putative virulence CDSs were identified.
Conclusion: S. uberis utilises nutritional flexibility derived from a diversity of metabolic options to successfully occupy a discrete ecological niche. The features observed in S. uberis are strongly suggestive of an opportunistic pathogen adapted to challenging and changing environmental parameters.
Figures
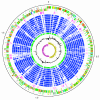
Figure 1
Schematic circular diagram of the S. uberis 0140J genome. Key for the circular diagram: scale (in Mb); annotated CDSs coloured according to predicted function represented on a pair of concentric circles, representing both coding strands; S. uberis unique CDSs, magenta; CDSs with Streptococcal ortholog matches, blue; ortholog matches shared with the Streptococcal species, S. pyogenes Manfredo, S. zooepidemicus H70, S. equi 4047, S. mutans UA159, S. gordonii Challis CH1, S. sanguinis SK36, S. pneumoniae TIGR4, S. agalactiae NEM316, S. suis P1/7, S. thermophilus CNRZ1066; Lactococcus lactis subsp. lactis, green; G + C% content plot; G + C deviation plot (>0% olive, <0% purple). Colour coding for CDS functions: dark blue; pathogenicity/adaptation, black; energy metabolism, red; information transfer, dark green; surface associated, cyan; degradation of large molecules, magenta; degradation of small molecules, yellow; central/intermediary metabolism, pale green; unknown, pale blue; regulators, orange; conserved hypothetical, brown; pseudogenes, pink; phage and IS elements, grey; miscellaneous.
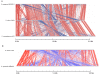
Figure 2
Genome comparison of pyogenic streptococci. Pairwise comparisons of the chromosomes of S. pyogenes MGAS315, S. uberis 0140J and S. zooepidemicus H70 (A), and S. uberis 0140J and agalactiae NEM316 (B) displayed using the Artemis Comparison Tool (ACT) [89]. The sequences have been aligned from the predicted replication origins (oriC; right). The coloured bars separating each genome (red and blue) represent similarity matches identified by reciprocal TBLASTX analysis [81], with a score cut off of 100. Red lines link matches in the same orientation; blue lines link matches in the reverse orientation.
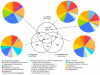
Figure 3
Distribution of orthologous CDSs in S. uberis strain 0140J, S. pyogenes strain Manfredo and S. zooepidemicus strain H70. The Venn diagram shows the number of predicted CDS unique or shared between one or more Streptococcal species. The figure in brackets indicates the number of unique CDSs that do not have ortholog matches in any of the other sequenced Streptococcus species. The relative distribution of functional groups of S. uberis 0140J CDSs in the various sections is illustrated in the pie charts. Colour legend for the functional classification is below.
Figure 4
Phylogenetic relationships of streptococcal glycoside hydrolase family 1 proteins. Maximum-likelihood tree of streptococcal glycoside hydrolase family 1 proteins from: S. uberis, dark blue; S. pyogenes Manfredo, red; S. equi 4047, brown; S. zooepidemicus H70 grey; S. suis P1/7, pink; S. pneumoniae TIGR4, S. gordonii Challis, green; light blue; S. sanguinis SK36, yellow; S. mutans UA159, purple; S. algalactiae NEM316, orange; black. Sequences of Fasta searches 'top match' hits in UniProt (Table 1), outside the streptococcal genomes used above have been included, black: Q3Y0U6_ENTFC, Enterococcus faecium; A1JLK3_YERE8, Yersinia enterocolitica; A5A692_BACLD, Bacillus licheniformis; Q300V6_STRSU, Streptococcus suis; Q65D52_BACLD, Bacillus licheniformis; Q3XXU1_ENTFC, Enterococcus faecium; LACG_LACLA, Lactococcus lactis subsp. lactis; Q02YI9_LACLS, Lactococcus lactis subsp. cremoris; A6M0G4_CLOB8, Clostridium beijerinckii. The tree was constructed with the WAG model of amino acid substitution, assuming a gamma distribution of among-site substitution rates. The numbers at the tree branches are percentage bootstrap values indicating the confidence levels, values below 50 are not shown.
Similar articles
- Virulence related sequences; insights provided by comparative genomics of Streptococcus uberis of differing virulence.
Hossain M, Egan SA, Coffey T, Ward PN, Wilson R, Leigh JA, Emes RD. Hossain M, et al. BMC Genomics. 2015 Apr 23;16(1):334. doi: 10.1186/s12864-015-1512-6. BMC Genomics. 2015. PMID: 25898893 Free PMC article. - Molecular characterization of Streptococcus agalactiae and Streptococcus uberis isolates from bovine milk.
Shome BR, Bhuvana M, Mitra SD, Krithiga N, Shome R, Velu D, Banerjee A, Barbuddhe SB, Prabhudas K, Rahman H. Shome BR, et al. Trop Anim Health Prod. 2012 Dec;44(8):1981-92. doi: 10.1007/s11250-012-0167-4. Epub 2012 May 16. Trop Anim Health Prod. 2012. PMID: 22588571 - Gene content differences across strains of Streptococcus uberis identified using oligonucleotide microarray comparative genomic hybridization.
Lang P, Lefébure T, Wang W, Zadoks RN, Schukken Y, Stanhope MJ. Lang P, et al. Infect Genet Evol. 2009 Mar;9(2):179-88. doi: 10.1016/j.meegid.2008.10.015. Epub 2008 Nov 14. Infect Genet Evol. 2009. PMID: 19056519 - The exploitation of the genome in the search for determinants of virulence in Streptococcus uberis.
Leigh JA, Ward PN, Field TR. Leigh JA, et al. Vet Immunol Immunopathol. 2004 Aug;100(3-4):145-9. doi: 10.1016/j.vetimm.2004.04.004. Vet Immunol Immunopathol. 2004. PMID: 15207452 Review. - Genomics, evolution, and molecular epidemiology of the Streptococcus bovis/Streptococcus equinus complex (SBSEC).
Jans C, Meile L, Lacroix C, Stevens MJ. Jans C, et al. Infect Genet Evol. 2015 Jul;33:419-36. doi: 10.1016/j.meegid.2014.09.017. Epub 2014 Sep 16. Infect Genet Evol. 2015. PMID: 25233845 Review.
Cited by
- Prediction of Streptococcus uberis clinical mastitis treatment success in dairy herds by means of mass spectrometry and machine-learning.
Maciel-Guerra A, Esener N, Giebel K, Lea D, Green MJ, Bradley AJ, Dottorini T. Maciel-Guerra A, et al. Sci Rep. 2021 Apr 8;11(1):7736. doi: 10.1038/s41598-021-87300-0. Sci Rep. 2021. PMID: 33833319 Free PMC article. - Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae.
Fernandez A, Lechardeur D, Derré-Bobillot A, Couvé E, Gaudu P, Gruss A. Fernandez A, et al. PLoS Pathog. 2010 Apr 22;6(4):e1000860. doi: 10.1371/journal.ppat.1000860. PLoS Pathog. 2010. PMID: 20421944 Free PMC article. - Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis.
Holden MT, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, Goodhead I, Mungall K, Quail MA, Price C, Rabbinowitsch E, Sharp S, Croucher NJ, Chieu TB, Mai NT, Diep TS, Chinh NT, Kehoe M, Leigh JA, Ward PN, Dowson CG, Whatmore AM, Chanter N, Iversen P, Gottschalk M, Slater JD, Smith HE, Spratt BG, Xu J, Ye C, Bentley S, Barrell BG, Schultsz C, Maskell DJ, Parkhill J. Holden MT, et al. PLoS One. 2009 Jul 15;4(7):e6072. doi: 10.1371/journal.pone.0006072. PLoS One. 2009. PMID: 19603075 Free PMC article. - Uptake of biotin by Chlamydia Spp. through the use of a bacterial transporter (BioY) and a host-cell transporter (SMVT).
Fisher DJ, Fernández RE, Adams NE, Maurelli AT. Fisher DJ, et al. PLoS One. 2012;7(9):e46052. doi: 10.1371/journal.pone.0046052. Epub 2012 Sep 27. PLoS One. 2012. PMID: 23029384 Free PMC article. - Discrimination of contagious and environmental strains of Streptococcus uberis in dairy herds by means of mass spectrometry and machine-learning.
Esener N, Green MJ, Emes RD, Jowett B, Davies PL, Bradley AJ, Dottorini T. Esener N, et al. Sci Rep. 2018 Nov 30;8(1):17517. doi: 10.1038/s41598-018-35867-6. Sci Rep. 2018. PMID: 30504894 Free PMC article.
References
- Bentley RW, Leigh JA, Collins MD. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int J Syst Bacteriol. 1991;41:487–494. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- S19514/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- S19514/3/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources