Structural determinants of integrin binding to the talin rod - PubMed (original) (raw)
. 2009 Mar 27;284(13):8866-76.
doi: 10.1074/jbc.M805937200. Epub 2009 Jan 27.
Wolfgang H Ziegler, Andrey A Bobkov, M Gordon Joyce, Domenico Fasci, Mirko Himmel, Sven Rothemund, Anett Ritter, J Günter Grossmann, Bipin Patel, Neil Bate, Benjamin T Goult, Jonas Emsley, Igor L Barsukov, Gordon C K Roberts, Robert C Liddington, Mark H Ginsberg, David R Critchley
Affiliations
- PMID: 19176533
- PMCID: PMC2659244
- DOI: 10.1074/jbc.M805937200
Structural determinants of integrin binding to the talin rod
Alexandre R Gingras et al. J Biol Chem. 2009.
Abstract
The adaptor protein talin serves both to activate the integrin family of cell adhesion molecules and to couple integrins to the actin cytoskeleton. Integrin activation has been shown to involve binding of the talin FERM domain to membrane proximal sequences in the cytoplasmic domain of the integrin beta-subunit. However, a second integrin-binding site (IBS2) has been identified near the C-terminal end of the talin rod. Here we report the crystal structure of IBS2 (residues 1974-2293), which comprises two five-helix bundles, "IBS2-A" (1974-2139) and "IBS2-B" (2140-2293), connected by a continuous helix with a distinct kink at its center that is stabilized by side-chain H-bonding. Solution studies using small angle x-ray scattering and NMR point to a fairly flexible quaternary organization. Using pull-down and enzyme-linked immunosorbent assays, we demonstrate that integrin binding requires both IBS2 domains, as does binding to acidic phospholipids and robust targeting to focal adhesions. We have defined the membrane proximal region of the integrin cytoplasmic domain as the major binding region, although more membrane distal regions are also required for strong binding. Alanine-scanning mutagenesis points to an important electrostatic component to binding. Thermal unfolding experiments show that integrin binding induces conformational changes in the IBS2 module, which we speculate are linked to vinculin and membrane binding.
Figures
FIGURE 1.
Domain structure and binding partners of talin. Schematic diagram of the talin molecule indicating the regions involved in binding to various ligands. The talin head (residues 1-400) contains a FERM domain (comprising_F1, F2_, and F3 subdomains) preceded by a domain referred to here as F0. The rod domain contains 62 predicted α-helices (ovals) organized into a series of amphipathic helical bundles. Domain boundaries based on structural determination are indicated by solid lines. Dashed lines indicate boundaries that are tentative. The ∼11 vinculin-binding sites (VBS) are shown in red. The last α-helix contains the dimerization domain (DD).
FIGURE 2.
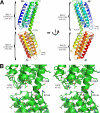
Structure of IBS2 in the talin rod. A, schematic representation of the talin 1974-2293 crystal structure. The upper five-helix bundle is called IBS2-A, and the lower one IBS2-B. The helix numbers shown in_brackets_ are for full-length talin. B, stereo representation of the area located between the two domains in the crystal structure; there is no evidence of hydrophobic or electrostatic interactions between the two domains.
FIGURE 3.
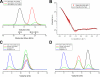
Biochemical characterization of the talin IBS2 polypeptide. A, talin polypeptides spanning residues 1974-2293 (IBS2), 1974-2140 (IBS2-A), and 2137-2293 (IBS2-B) were analyzed on a Superdex-75 (10/300) GL gel filtration column. The apparent molecular mass for each domain is indicated with their theoretical molecular mass in brackets. The talin IBS2 polypeptides showed an anomalous elution profile indicative of an extended conformation. B, SAXS of the talin IBS2 polypeptide indicates a different domain organization from that in the crystal structure. Experimental scattering profile of talin IBS2 (red) compared with the simulated scattering profile based on the crystal structure (black line) (goodness-of-fitχ = 8.9). C and D, binding of the vinculin Vd1 domain to talin IBS2 (C) or IBS2-A (D) was analyzed on a Superdex-75 (10/300) GL gel filtration column at room temperature (RT). Incubation of either IBS2 or IBS2-A with Vd1 at room temperature resulted in rather little complex formation, and most of the talin and vinculin polypeptides remained in the free form. However, preincubation of the proteins at 37 °C resulted in formation of a talin-Vd1 complex.
FIGURE 4.
The talin 1974-2293 IBS2 polypeptide binds to β3-integrin tails in pulldown and ELISA-type assays. A, schematic of talin rod polypeptides used where each box represents a five-helix bundle with the exception of the C-terminal dimerization domain, which is composed of a single α-helix that forms an anti-parallel dimer (16). All constructs include an N-terminal His tag followed by a V5 epitope. B, pulldown assays using αIIb- and β3-integrin tails immobilized on NeutrAvidin beads with purified recombinant talin rod polypeptides. Binding of talin rod polypeptides was detected using an anti-V5 antibody, while binding of the talin head (used as a positive control) was detected using an anti-His antibody (data not shown). C and D, binding of talin polypeptides to microtiter wells coated with β3-integrins using ELISA. The talin head polypeptide (residues 1-405) was used as a positive control. Binding to wells coated with αIIb-integrin or not coated with integrins were used as negative controls. The talin IBS2 polypeptide, which contains both the IBS2-A and IBS2-B five-helix bundles, binds to β3-integrin with much higher affinity than the individual IBS2-A and IBS2-B five-helix bundles.
FIGURE 5.
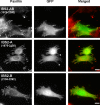
GFP-talin IBS2 localizes to FAs in vinculin null cells. Mouse embryonic fibroblasts derived from vinculin knockout mice (39) were transfected with cDNAs encoding EGFP-tagged IBS2 fragments. FAs were visualized by paxillin staining. The IBS2 double domain construct clearly localized to FAs, whereas the individual five-helix bundles IBS2-A and IBS2-B showed little or no targeting.
FIGURE 6.
Talin IBS2 but not IBS2-A or IBS2-B binds to acidic phospholipids. Binding of His-tagged talin polypeptides to phosphatidylinositol phosphate strips containing an array of acidic phospholipids (Invitrogen) was detected using an anti-His antibody. Each spot contains 100 pmol of phospholipid, and the membrane was challenged with 1 μg/ml protein. Talin IBS2 (residues 1974-2293) binds to several phospholipids, whereas the individual five-helix bundles that make up IBS2, i.e. residues 1974-2140 (IBS2-A) and residues 2137-2293 (IBS2-B), did not bind to any of the phospholipids tested.
FIGURE 7.
Talin IBS2 binds to membrane proximal β-integrin tail peptides. A, alignment of the full-length β1A-integrin cytoplasmic domain peptide with the membrane proximal, middle, and distal tail peptides used in this study. The membrane proximal helical region and NP_X_Y motif are underlined. Two schematic representations of the β-integrin cytoplasmic domain highlight key positions. The amino acid numbering is for β1A-integrin. B and C, analysis of the binding of a talin 1975-2541 polypeptide to a series of immobilized β-integrin cytoplasmic domain peptides. B, binding to membrane proximal (top), middle, and distal tail peptides equivalent to β1A-, β2-, β3-, and β7-integrins (25-mers). Binding to membrane proximal peptides from all β-integrin tails was observed (including β5 and β6; data not shown). C, analysis of binding to a series of membrane proximal β-integrin peptides (25-mers) in which each residue in turn was substituted by alanine. Amino acid substitutions that consistently affected talin binding are_highlighted_. Amino acid numbering for β1A-integrin is shown. Mutation of β-integrin membrane proximal peptides identify important charged residues required for optimal talin 1975-2541 binding (see_A_).
FIGURE 8.
DSC analysis shows binding of various talin rod domains to β-integrin tail peptides in solution. DSC analysis of talin (A and D) 1974-2293, (B and E) 1974-2140 (IBS2-A), and (C and F) 2137-2293 (IBS2-B) in the presence of 47-mer β1A-integrin peptide (A-C) and 25-mer β1A-integrin membrane proximal peptide (D-F). The concentration of the talin constructs was 0.7 mg/ml.
Similar articles
- Crystal structure of vinculin in complex with vinculin binding site 50 (VBS50), the integrin binding site 2 (IBS2) of talin.
Yogesha SD, Rangarajan ES, Vonrhein C, Bricogne G, Izard T. Yogesha SD, et al. Protein Sci. 2012 Apr;21(4):583-8. doi: 10.1002/pro.2041. Epub 2012 Feb 28. Protein Sci. 2012. PMID: 22334306 Free PMC article. - The talin rod IBS2 alpha-helix interacts with the beta3 integrin cytoplasmic tail membrane-proximal helix by establishing charge complementary salt bridges.
Rodius S, Chaloin O, Moes M, Schaffner-Reckinger E, Landrieu I, Lippens G, Lin M, Zhang J, Kieffer N. Rodius S, et al. J Biol Chem. 2008 Aug 29;283(35):24212-23. doi: 10.1074/jbc.M709704200. Epub 2008 Jun 23. J Biol Chem. 2008. PMID: 18577523 Free PMC article. - The integrin binding site 2 (IBS2) in the talin rod domain is essential for linking integrin beta subunits to the cytoskeleton.
Moes M, Rodius S, Coleman SJ, Monkley SJ, Goormaghtigh E, Tremuth L, Kox C, van der Holst PP, Critchley DR, Kieffer N. Moes M, et al. J Biol Chem. 2007 Jun 8;282(23):17280-8. doi: 10.1074/jbc.M611846200. Epub 2007 Apr 11. J Biol Chem. 2007. PMID: 17430904 - Integrin connections to the cytoskeleton through talin and vinculin.
Ziegler WH, Gingras AR, Critchley DR, Emsley J. Ziegler WH, et al. Biochem Soc Trans. 2008 Apr;36(Pt 2):235-9. doi: 10.1042/BST0360235. Biochem Soc Trans. 2008. PMID: 18363566 Review. - Integrin-mediated cell adhesion: the cytoskeletal connection.
Critchley DR, Holt MR, Barry ST, Priddle H, Hemmings L, Norman J. Critchley DR, et al. Biochem Soc Symp. 1999;65:79-99. Biochem Soc Symp. 1999. PMID: 10320934 Review.
Cited by
- High-resolution snapshots of the talin auto-inhibitory states suggest roles in cell adhesion and signaling.
Rangarajan ES, Bois JL, Hansen SB, Izard T. Rangarajan ES, et al. Nat Commun. 2024 Oct 28;15(1):9270. doi: 10.1038/s41467-024-52581-2. Nat Commun. 2024. PMID: 39468080 Free PMC article. - Talin-1 variants associated with spontaneous coronary artery dissection (SCAD) highlight how even subtle changes in multi-functional scaffold proteins can manifest in disease.
Azizi L, Otani Y, Mykuliak VV, Goult BT, Hytönen VP, Turkki P. Azizi L, et al. Hum Mol Genet. 2024 Nov 5;33(21):1846-1857. doi: 10.1093/hmg/ddae120. Hum Mol Genet. 2024. PMID: 39163585 Free PMC article. - Intrinsic self-organization of integrin nanoclusters within focal adhesions is required for cellular mechanotransduction.
Jain K, Lim KYE, Sheetz MP, Kanchanawong P, Changede R. Jain K, et al. bioRxiv [Preprint]. 2023 Nov 20:2023.11.20.567975. doi: 10.1101/2023.11.20.567975. bioRxiv. 2023. PMID: 38045378 Free PMC article. Preprint. - Calpain Promotes LPS-induced Lung Endothelial Barrier Dysfunction via Cleavage of Talin.
Song L, Shi X, Kovacs L, Han W, John J, Barman SA, Dong Z, Lucas R, Fulton DJR, Verin AD, Su Y. Song L, et al. Am J Respir Cell Mol Biol. 2023 Dec;69(6):678-688. doi: 10.1165/rcmb.2023-0009OC. Am J Respir Cell Mol Biol. 2023. PMID: 37639326 Free PMC article. - The structural basis of β2 integrin intra-cellular multi-protein complexes.
Bhattacharjya S. Bhattacharjya S. Biophys Rev. 2022 Sep 7;14(5):1183-1195. doi: 10.1007/s12551-022-00995-x. eCollection 2022 Oct. Biophys Rev. 2022. PMID: 36345283 Free PMC article. Review.
References
- Critchley, D. R., and Gingras, A. R. (2008) J. Cell Sci. 121 1345-1347 - PubMed
- McLachlan, A. D., Stewart, M., Hynes, R. O., and Rees, D. J. (1994) J. Mol. Biol. 235 1278-1290 - PubMed
- Gingras, A. R., Ziegler, W. H., Frank, R., Barsukov, I. L., Roberts, G. C., Critchley, D. R., and Emsley, J. (2005) J. Biol. Chem. 280 37217-37224 - PubMed
- Garcia-Alvarez, B., de Pereda, J. M., Calderwood, D. A., Ulmer, T. S., Critchley, D., Campbell, I. D., Ginsberg, M. H., and Liddington, R. C. (2003) Mol. Cell 11 49-58 - PubMed
- Wegener, K. L., Partridge, A. W., Han, J., Pickford, A. R., Liddington, R. C., Ginsberg, M. H., and Campbell, I. D. (2007) Cell 128 171-182 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources