Splice-site pairing is an intrinsically high fidelity process - PubMed (original) (raw)
Splice-site pairing is an intrinsically high fidelity process
Kristi L Fox-Walsh et al. Proc Natl Acad Sci U S A. 2009.
Abstract
The extensive alternative splicing in higher eukaryotes has initiated a debate whether alternative mRNA isoforms are generated by an inaccurate spliceosome or are the consequence of highly degenerate splice sites within the human genome. Here, we established a quantitative assay to evaluate the accuracy of splice-site pairing by determining the number of incorrect exon-skipping events made from constitutively spliced pre-mRNA transcripts. We demonstrate that the spliceosome pairs exons with an astonishingly high degree of accuracy that may be limited by the quality of pre-mRNAs generated by RNA pol II. The error rate of exon pairing is increased by the effects of the neurodegenerative disorder spinal muscular atrophy because of reduced levels of Survival of Motor Neuron, a master assembler of spliceosomal components. We conclude that all multi-intron-containing genes are alternatively spliced and that the reduction of SMN results in a general splicing defect that is mediated through alterations in the fidelity of splice-site pairing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
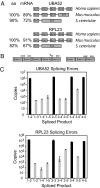
Error rates of pre-mRNA splicing. (A) The exon/intron structures and phylogenetic conservation [amino acid (aa) and mRNA] of UBA52 and RPL23 are shown. (B) Experimental design using exon junction primers amplifying all mRNA isoforms generated through alternative exon ligation. Correct priming of the 1–2 splice variant is shown on the left. The middle set of primers shows incorrect priming of the 2–5 exon junction primer. The primers on the far right show correct priming of the 2–5 exon junction primer set. (C) Bar graph showing the total copy number detected for every exon skipping (gray) and constitutive splicing (black) event for all possible exon junctions in UBA52 and RPL23. cDNAs used to determine the error rate were generated with a mixture of random hexamers. Error rates are summarized in Table 1.
Fig. 2.
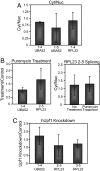
Inhibition of translation and NMD does not affect the error rate of splicing. (A) Error analysis was performed and compared between RNA samples isolated from cytoplasmic and nuclear fractions. No statistically significant difference in the error rate is detected between nuclear and cytoplasmic samples, implying uniform export and half-life kinetics between mRNA isoforms tested. Amplification of nuclear U6 snRNA was used to confirm efficient nuclear and cytoplasmic fractionation (
Fig. S4
). (B) Comparison of splicing errors in the presence or absence of the translation inhibitor puromycin. Error analysis was performed without (left panel) or with (right panel) nuclear/cytoplasmic fractionation. (C) The effect of hUpf1 knockdown on the error rate of splicing. hUpf1 knockdown was confirmed by RT-PCR, Western Blot, and splicing analysis of NMD target genes that are differentially spliced upon hUpf1 knockdown (
Fig. S5
). *, P < 0.05
Fig. 3.
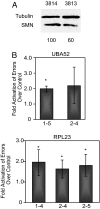
SMA patient fibroblast cells show an increase in the error rate of splicing. (A) Western blot highlighting the difference in SMN levels between SMA patient (3813) and control (3814) cells. The quantitation shown below was normalized to tubulin levels. (B) Bar graph illustrating the relative increase of the splicing error rate in SMA patient cells. *, P < 0.05
Fig. 4.
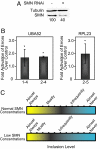
Decreased SMN concentrations increase the error rate of splicing. (A) Western blot confirming SMN knockdown in HeLa cells. The quantitation shown below was normalized to tubulin levels. SMN knockdown is also confirmed by RT-PCR (
Fig. S7
). (B) Bar graph showing the relative increase in splicing errors when cells are treated with RNAi against SMN. (C) Model showing the effect of normal versus reduced SMN concentrations on the recognition and inclusion of exons. At normal concentrations the scale of recognition is shown (Upper). When SMN concentrations are reduced, some alternatively spliced exons are less frequently included (Lower). *, P < 0.05.
Similar articles
- Ultra-deep sequencing reveals pre-mRNA splicing as a sequence driven high-fidelity process.
Reynolds DJ, Hertel KJ. Reynolds DJ, et al. PLoS One. 2019 Oct 3;14(10):e0223132. doi: 10.1371/journal.pone.0223132. eCollection 2019. PLoS One. 2019. PMID: 31581208 Free PMC article. - The architecture of pre-mRNAs affects mechanisms of splice-site pairing.
Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ. Fox-Walsh KL, et al. Proc Natl Acad Sci U S A. 2005 Nov 8;102(45):16176-81. doi: 10.1073/pnas.0508489102. Epub 2005 Oct 31. Proc Natl Acad Sci U S A. 2005. PMID: 16260721 Free PMC article. - Mechanism of Splicing Regulation of Spinal Muscular Atrophy Genes.
Singh RN, Singh NN. Singh RN, et al. Adv Neurobiol. 2018;20:31-61. doi: 10.1007/978-3-319-89689-2_2. Adv Neurobiol. 2018. PMID: 29916015 Free PMC article. Review. - Activation of a cryptic 5' splice site reverses the impact of pathogenic splice site mutations in the spinal muscular atrophy gene.
Singh NN, Del Rio-Malewski JB, Luo D, Ottesen EW, Howell MD, Singh RN. Singh NN, et al. Nucleic Acids Res. 2017 Dec 1;45(21):12214-12240. doi: 10.1093/nar/gkx824. Nucleic Acids Res. 2017. PMID: 28981879 Free PMC article. - Cancer-Associated Perturbations in Alternative Pre-messenger RNA Splicing.
Shkreta L, Bell B, Revil T, Venables JP, Prinos P, Elela SA, Chabot B. Shkreta L, et al. Cancer Treat Res. 2013;158:41-94. doi: 10.1007/978-3-642-31659-3_3. Cancer Treat Res. 2013. PMID: 24222354 Review.
Cited by
- Stoichiometry of a regulatory splicing complex revealed by single-molecule analyses.
Cherny D, Gooding C, Eperon GE, Coelho MB, Bagshaw CR, Smith CW, Eperon IC. Cherny D, et al. EMBO J. 2010 Jul 7;29(13):2161-72. doi: 10.1038/emboj.2010.103. Epub 2010 May 25. EMBO J. 2010. PMID: 20502437 Free PMC article. - Epigenetics in alternative pre-mRNA splicing.
Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Luco RF, et al. Cell. 2011 Jan 7;144(1):16-26. doi: 10.1016/j.cell.2010.11.056. Cell. 2011. PMID: 21215366 Free PMC article. Review. - Illuminating the Transcriptome through the Genome.
Elliott DJ. Elliott DJ. Genes (Basel). 2014 Mar 14;5(1):235-53. doi: 10.3390/genes5010235. Genes (Basel). 2014. PMID: 24705295 Free PMC article. - Re-evaluating the impact of alternative RNA splicing on proteomic diversity.
Manuel JM, Guilloy N, Khatir I, Roucou X, Laurent B. Manuel JM, et al. Front Genet. 2023 Feb 9;14:1089053. doi: 10.3389/fgene.2023.1089053. eCollection 2023. Front Genet. 2023. PMID: 36845399 Free PMC article. Review. - Quantitative evaluation of all hexamers as exonic splicing elements.
Ke S, Shang S, Kalachikov SM, Morozova I, Yu L, Russo JJ, Ju J, Chasin LA. Ke S, et al. Genome Res. 2011 Aug;21(8):1360-74. doi: 10.1101/gr.119628.110. Epub 2011 Jun 9. Genome Res. 2011. PMID: 21659425 Free PMC article.
References
- Burge CB, Tuschl T, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2nd Ed. Cold Spring Harbor, New York: CSHL Press; 1999. pp. 525–560.
- Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–546. - PubMed
- Chua K, Reed R. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature. 1999;402:207–210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources