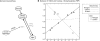Coordinated concentration changes of transcripts and metabolites in Saccharomyces cerevisiae - PubMed (original) (raw)
Coordinated concentration changes of transcripts and metabolites in Saccharomyces cerevisiae
Patrick H Bradley et al. PLoS Comput Biol. 2009 Jan.
Abstract
Metabolite concentrations can regulate gene expression, which can in turn regulate metabolic activity. The extent to which functionally related transcripts and metabolites show similar patterns of concentration changes, however, remains unestablished. We measure and analyze the metabolomic and transcriptional responses of Saccharomyces cerevisiae to carbon and nitrogen starvation. Our analysis demonstrates that transcripts and metabolites show coordinated response dynamics. Furthermore, metabolites and gene products whose concentration profiles are alike tend to participate in related biological processes. To identify specific, functionally related genes and metabolites, we develop an approach based on Bayesian integration of the joint metabolomic and transcriptomic data. This algorithm finds interactions by evaluating transcript-metabolite correlations in light of the experimental context in which they occur and the class of metabolite involved. It effectively predicts known enzymatic and regulatory relationships, including a gene-metabolite interaction central to the glycolytic-gluconeogenetic switch. This work provides quantitative evidence that functionally related metabolites and transcripts show coherent patterns of behavior on the genome scale and lays the groundwork for building gene-metabolite interaction networks directly from systems-level data.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
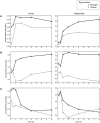
Figure 1. Singular value decomposition reveals coordination between transcriptional and metabolic responses to carbon and nitrogen starvation.
Eigenvectors of the transcription (left) and metabolite (right) concentration data sets were calculated. Each eigenvector is composed of a characteristic response under carbon starvation (“carbon”, in light gray circles) and under nitrogen starvation (“nitrogen”, in dark gray triangles). For each gene eigenvector on the left, the corresponding metabolite eigenvector is plotted on the right. The corresponding eigenvectors correlated significantly, with p-values of (A) 6.9×10−3, (B) 2.2×10−4, and (C) 2.1×10−2. For the transcript data, the percent information explained by each eigenvector was (A) 46%, (B) 13%, and (C) 9%. For the metabolite data, the percent information explained was (A) 33%, (B) 20%, and (C) 10%.
Figure 2. A selection of example scatterplots that demonstrate experimental condition-dependent correlation between metabolites and related genes, motivating the use of a Bayesian algorithm.
Metabolite and gene transcript concentration changes are represented as  ratios of measurements from starved cells to measurements from unstarved cells. The responses observed under carbon starvation (light gray circles, “Carbon” in legend) are labeled distinctly from the responses under nitrogen starvation (dark gray triangles, “Nitrogen” in legend), but are plotted on the same axes. Solid light gray and dark gray lines are linear best-fits for the responses observed under carbon and nitrogen starvation, respectively; the dashed line is a linear best-fit curve for all data. (A–E) Scatterplots of metabolites from the glycolysis and pentose-phosphate pathway metabolic class versus related genes show an inverse relationship under carbon starvation, but a positive correlation under nitrogen starvation. The dashed line shows that this relationship would be obscured by computing correlation across all data points. ILV2 catalyzes the first step of isoleucine and valine biosynthesis from pyruvate; ARO3 catalyzes the first step in aromatic amino acid biosynthesis from PEP and erythrose-4-phosphate; ALD6, which also plays a key role in redox metabolism, is involved in the creation of cytosolic acetyl-CoA from pyruvate; GLK1 phosphorylates glucose to glucose-6-phosphate; and PGM2 catalyzes the interconversion of glucose-1-phosphate and glucose-6-phosphate. (F–H) Scatterplots of metabolites from the amino acid metabolic class versus related genes, in contrast, show positive correlation in both carbon and nitrogen starvation. Even in this case, however, computing correlation across both conditions can lead to an underestimation of the extent of the relationship (e.g., (H) threonine vs. THR4, where although
ratios of measurements from starved cells to measurements from unstarved cells. The responses observed under carbon starvation (light gray circles, “Carbon” in legend) are labeled distinctly from the responses under nitrogen starvation (dark gray triangles, “Nitrogen” in legend), but are plotted on the same axes. Solid light gray and dark gray lines are linear best-fits for the responses observed under carbon and nitrogen starvation, respectively; the dashed line is a linear best-fit curve for all data. (A–E) Scatterplots of metabolites from the glycolysis and pentose-phosphate pathway metabolic class versus related genes show an inverse relationship under carbon starvation, but a positive correlation under nitrogen starvation. The dashed line shows that this relationship would be obscured by computing correlation across all data points. ILV2 catalyzes the first step of isoleucine and valine biosynthesis from pyruvate; ARO3 catalyzes the first step in aromatic amino acid biosynthesis from PEP and erythrose-4-phosphate; ALD6, which also plays a key role in redox metabolism, is involved in the creation of cytosolic acetyl-CoA from pyruvate; GLK1 phosphorylates glucose to glucose-6-phosphate; and PGM2 catalyzes the interconversion of glucose-1-phosphate and glucose-6-phosphate. (F–H) Scatterplots of metabolites from the amino acid metabolic class versus related genes, in contrast, show positive correlation in both carbon and nitrogen starvation. Even in this case, however, computing correlation across both conditions can lead to an underestimation of the extent of the relationship (e.g., (H) threonine vs. THR4, where although  and
and  ,
,  ). HTS1 charges (i.e. aminoacylates) the histidinyl-tRNA; MET6 catalyzes the formation of methionine from homocysteine; and THR4 converts phosphohomoserine to threonine.
). HTS1 charges (i.e. aminoacylates) the histidinyl-tRNA; MET6 catalyzes the formation of methionine from homocysteine; and THR4 converts phosphohomoserine to threonine.
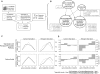
Figure 3. Bayesian network relating gene–metabolite interactions to metabolomic and transcriptomic data and to metabolite class.
(A) Overview of Bayesian integration procedure. Transcript and metabolite data were used to compute correlations between genes and metabolites over time under different experimental conditions. These correlations, along with a set of positive and negative examples obtained from KEGG, were used to train a Bayesian network. (B) Structure of the Bayesian network. This four-node network states that the variables corresponding to gene–metabolite correlations observed under either nitrogen or carbon starvation depend on the class of the metabolite involved, and whether or not a functional relationship between the gene and metabolite exists. The rounded boxes by each node represent the possible values that the nodes can take. (C) Conditional probability distributions learned from the experimental data. The parameters of the Bayesian network were computed from the experimental data and the set of positive and negative examples of gene–metabolite functional interactions. The light gray line gives the probability (y-axis) that, given no functional relationship, one would observe a given correlation (x-axis); the dark gray line gives the corresponding probability if given a true functional relationship instead. (D) Conditional probability distributions represented as log-odds scores. The sign of the bar corresponds to whether observing a certain strength and direction of correlation is more likely for a true functional relationship (positive) or for no functional relationship (negative), while the magnitude of the bar corresponds to how much more likely this is.
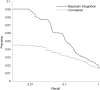
Figure 4. Precision-recall curve (PRC) showing the superior performance of context-sensitive Bayesian integration (dark gray line), as compared to overall strength of gene–metabolite concentration correlation (light gray line), for identifying gene–metabolite functional interactions from transcriptomic and metabolomic data.
Recall, or fraction of known positives predicted by the system, is plotted on the X-axis (log scale); precision, or fraction of predictions that are in the training set, is plotted on the Y axis. The Bayesian integration PRC shows greater area under the curve than the correlation PRC, especially in the left-most, highest-confidence regime.
Figure 5. Example of gene regulating the glycolytic–gluconeogenic switch (VID24) that we identified as interacting with the key glycolytic metabolite fructose-1,6-bisphosphate (FBP).
(A) Schematic of reactions involving FBP and VID24. The conversion of FBP to hexose phosphate is catalyzed by fructose-1,6-bisphosphatase (FBP1). Vid24p destroys this enzyme by targeting it to the vacuole for destruction. (B) Scatterplot showing the relationship between VID24 and FBP abundances over carbon starvation (“carbon” in the legend) and nitrogen starvation (“nitrogen” in the legend). As in Figure 2, lines represent linear best-fit curves, calculated separately for each condition (solid lines) or over both conditions (dashed line). VID24 and FBP are inversely correlated under carbon starvation (light gray), but positively correlated under nitrogen starvation (dark gray), as anticipated for a gene interacting with a glycolytic metabolite. VID24 was in the top 3% of predictions made for FBP.
Similar articles
- Quantitative Metabolomics of Saccharomyces Cerevisiae Using Liquid Chromatography Coupled with Tandem Mass Spectrometry.
Mohammad K, Jiang H, Titorenko VI. Mohammad K, et al. J Vis Exp. 2021 Jan 5;(167). doi: 10.3791/62061. J Vis Exp. 2021. PMID: 33491678 - Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.
Wolahan SM, Hirt D, Glenn TC. Wolahan SM, et al. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review. - Targeted proteome analysis of single-gene deletion strains of Saccharomyces cerevisiae lacking enzymes in the central carbon metabolism.
Matsuda F, Kinoshita S, Nishino S, Tomita A, Shimizu H. Matsuda F, et al. PLoS One. 2017 Feb 27;12(2):e0172742. doi: 10.1371/journal.pone.0172742. eCollection 2017. PLoS One. 2017. PMID: 28241048 Free PMC article. - Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation.
Zhu J, Sova P, Xu Q, Dombek KM, Xu EY, Vu H, Tu Z, Brem RB, Bumgarner RE, Schadt EE. Zhu J, et al. PLoS Biol. 2012;10(4):e1001301. doi: 10.1371/journal.pbio.1001301. Epub 2012 Apr 3. PLoS Biol. 2012. PMID: 22509135 Free PMC article. - The top genes: on the distance from transcript to function in yeast glycolysis.
Fraenkel DG. Fraenkel DG. Curr Opin Microbiol. 2003 Apr;6(2):198-201. doi: 10.1016/s1369-5274(03)00023-7. Curr Opin Microbiol. 2003. PMID: 12732312 Review.
Cited by
- Metabolomic analysis of the selection response of Drosophila melanogaster to environmental stress: are there links to gene expression and phenotypic traits?
Malmendal A, Sørensen JG, Overgaard J, Holmstrup M, Nielsen NC, Loeschcke V. Malmendal A, et al. Naturwissenschaften. 2013 May;100(5):417-27. doi: 10.1007/s00114-013-1040-7. Epub 2013 Apr 10. Naturwissenschaften. 2013. PMID: 23571708 - The SAGA complex, together with transcription factors and the endocytic protein Rvs167p, coordinates the reprofiling of gene expression in response to changes in sterol composition in Saccharomyces cerevisiae.
Dewhurst-Maridor G, Abegg D, David FPA, Rougemont J, Scott CC, Adibekian A, Riezman H. Dewhurst-Maridor G, et al. Mol Biol Cell. 2017 Oct 1;28(20):2637-2649. doi: 10.1091/mbc.E17-03-0169. Epub 2017 Aug 2. Mol Biol Cell. 2017. PMID: 28768829 Free PMC article. - A novel connection between the Cell Wall Integrity and the PKA pathways regulates cell wall stress response in yeast.
García R, Bravo E, Diez-Muñiz S, Nombela C, Rodríguez-Peña JM, Arroyo J. García R, et al. Sci Rep. 2017 Jul 18;7(1):5703. doi: 10.1038/s41598-017-06001-9. Sci Rep. 2017. PMID: 28720901 Free PMC article. - Consensus-phenotype integration of transcriptomic and metabolomic data implies a role for metabolism in the chemosensitivity of tumour cells.
Cavill R, Kamburov A, Ellis JK, Athersuch TJ, Blagrove MS, Herwig R, Ebbels TM, Keun HC. Cavill R, et al. PLoS Comput Biol. 2011 Mar;7(3):e1001113. doi: 10.1371/journal.pcbi.1001113. Epub 2011 Mar 31. PLoS Comput Biol. 2011. PMID: 21483477 Free PMC article. - Yeast Cip1 is activated by environmental stress to inhibit Cdk1-G1 cyclins via Mcm1 and Msn2/4.
Chang YL, Tseng SF, Huang YC, Shen ZJ, Hsu PH, Hsieh MH, Yang CW, Tognetti S, Canal B, Subirana L, Wang CW, Chen HT, Lin CY, Posas F, Teng SC. Chang YL, et al. Nat Commun. 2017 Jul 4;8(1):56. doi: 10.1038/s41467-017-00080-y. Nat Commun. 2017. PMID: 28676626 Free PMC article.
References
- Sauer U, Heinemann M, Zamboni N. Genetics. Getting closer to the whole picture. Science. 2007;316:550–551. - PubMed
- Askenazi M, Driggers EM, Holtzman DA, Norman TC, Iverson S, et al. Integrating transcriptional and metabolite profiles to direct the engineering of lovastatin-producing fungal strains. Nat Biotechnol. 2003;21:150–156. - PubMed
- Suzuki H, Reddy MSS, Naoumkina M, Aziz N, May GD, et al. Methyl jasmonate and yeast elicitor induce differential transcriptional and metabolic re-programming in cell suspension cultures of the model legume Medicago truncatula. Planta. 2005;220:696–707. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases