Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization - PubMed (original) (raw)
. 2009 Apr 2;113(14):3314-22.
doi: 10.1182/blood-2008-04-154310. Epub 2009 Feb 2.
Laurie E Riesbeck, Chinavenmeni S Velu, Aditya Chaubey, Jiwang Zhang, Nicholas J Achille, Frank E Erfurth, Katherine Eaton, Jun Lu, H Leighton Grimes, Jianjun Chen, Janet D Rowley, Nancy J Zeleznik-Le
Affiliations
- PMID: 19188669
- PMCID: PMC2665896
- DOI: 10.1182/blood-2008-04-154310
Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization
Relja Popovic et al. Blood. 2009.
Abstract
Chromosomal translocations involving the Mixed Lineage Leukemia (MLL) gene produce chimeric proteins that cause abnormal expression of a subset of HOX genes and leukemia development. Here, we show that MLL normally regulates expression of mir-196b, a hematopoietic microRNA located within the HoxA cluster, in a pattern similar to that of the surrounding 5' Hox genes, Hoxa9 and Hoxa10, during embryonic stem (ES) cell differentiation. Within the hematopoietic lineage, mir-196b is most abundant in short-term hematopoietic stem cells and is down-regulated in more differentiated hematopoietic cells. Leukemogenic MLL fusion proteins cause overexpression of mir-196b, while treatment of MLL-AF9 transformed bone marrow cells with mir-196-specific antagomir abrogates their replating potential in methylcellulose. This demonstrates that mir-196b function is necessary for MLL fusion-mediated immortalization. Furthermore, overexpression of mir-196b was found specifically in patients with MLL associated leukemias as determined from analysis of 55 primary leukemia samples. Overexpression of mir-196b in bone marrow progenitor cells leads to increased proliferative capacity and survival, as well as a partial block in differentiation. Our results suggest a mechanism whereby increased expression of mir-196b by MLL fusion proteins significantly contributes to leukemia development.
Figures
Figure 1
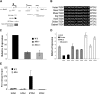
Expression of mir-196b is dependent on Mll and Menin. (A) Schematic illustration of the murine Hoxa9 locus on chromosomal band 6qB3. Open boxes represent 3 exons and gray boxes indicate CpG islands. Exon II is the homeodomain (HD) containing exon. The canonical Hoxa9 is encoded by exons CD and II. Regions of high homology between species are noted as dark boxes and the region amplified by ChIP primers is indicated by arrows. The location of mir-196b is labeled above exon AB with an arrowhead. (B) Sequence alignment of the conserved mir-196b sequence among different species. The mature mir-196b sequence is highlighted in black. (C) Quantitative RT-PCR of mir-196b levels in WT, _Mll_−/−, and _Men_−/− MEFs. The experiment was run in triplicate and the results represent average expression plus or minus SD. (D) Quantitative RT-PCR of mir-196b expression in individual clones expressing an empty vector, MLL, or MLLAF4 in _Mll_−/− MEF background. Transfection of _Mll_−/− MEFs and clone selection was performed as previously described. Mir-196b expression was determined from total RNA. (E) ChIP assay performed in WT and _Mll_−/− MEFs. Chromatin was precipitated using indicated antibodies and qPCR performed with primers spanning mir-196b region. All samples were run in triplicate and were normalized to the input chromatin.
Figure 2
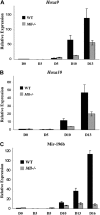
Mir-196b is regulated by Mll during ES cell differentiation, similar to Hoxa9 and Hoxa10. (A,B) Quantitative RT-PCR of Hoxa9 (A) and Hoxa10 (B) mRNA using SYBR green. All values are compared with WT day 0 and this value is set to 1. Experiments were performed in triplicate and the results represent average fold change plus or minus SD. (C) Quantitative RT-PCR of mir-196b expression using mir-196b TaqMan primers and probe.
Figure 3
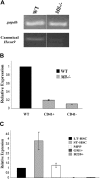
Mll regulates mir-196b expression in hematopoietic progenitors. (A) RT-PCR for canonical Hoxa9 in CD41+ ES cells. Day 10 EBs were disrupted and CD41+ cells isolated using a magnetic column. Total RNA was isolated from the CD41+ cells and used for cDNA synthesis. Hoxa9 primers used span the junction between exon CD and exon II of the gene. Gapdh is used as a loading control. Space between panels indicates repositioned gel lanes. (B) Quantitative RT-PCR of mir-196b expression in WT and _Mll_−/−CD41+ and CD41− murine embryonic stem cells from day 10 embryoid bodies. WT levels were set to 1 for comparison. Experiment was performed in triplicate and the results represent average expression plus or minus SD. (C) Quantitative RT-PCR of mir-196b expression in sorted mouse bone marrow cells. Mouse bone marrow cells were sorted in various populations based on the expression of cell surface markers. Expression of LT-HSCs is set to1 for comparison. The results represent average fold change plus or minus SD.
Figure 4
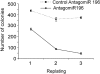
MLL fusion proteins induce expression of mir-196b in primary bone marrow progenitors, which is required for their MLL fusion-dependent increased proliferative capacity in vitro. Wild-type Lin− bone marrow cells were transduced with MLL-AF9 retroviral vectors and treated with either control antagomiR-196 or antagomiR-196, then plated in duplicate. Colonies were enumerated after 8 days and the average number was plotted. Cells were then replated after additional antagomir treatment. The cycle was repeated twice. Data shown are representative of 2 independent experiments with similar results.
Figure 5
Mir-196b is overexpressed in the majority of MLL-associated leukemias, but not in non-MLL leukemias, irrespective of their phenotype. Heat map of relative mir-196b expression of 55 leukemia samples and 3 normal controls using bead-based technology.
Figure 6
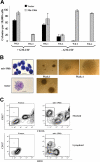
Mir-196b expression enhances colony formation and partially blocks hematopoietic progenitor cell differentiation. (A,B) Serial replating myeloid colony assay using c-Kit+ bone marrow cells. Cells were transduced with retroviruses producing mir-196b construct or with an empty vector and plated in methylcellulose with 2 different cytokine mixes. The number of colonies (A) and colony and cell morphology (B) at 1 and 4 weeks are shown. Only mir-196b–expressing cells are able to form colonies at the fourth week. Scale bars on cytospin pictures represent 10 μm in top panel and 25 μm in bottom panel. (C) Representative FACS profiles of in vitro–differentiated bone marrow cells. After 1 week of growth and selection in methylcellulose, mir-196b or vector-expressing bone marrow cells were plated on the OP9 cell line in the presence of GM-CSF or IL-7/Flt3L. After 5 days, expression of CD117 and CD11b (top panels, cells on GM-CSF) or B220 (bottom panels, cells on IL-7/Flt3L) was determined. Under both conditions, overexpression of mir-196b causes a partial block in differentiation.
Figure 7
Model for the role of mir-196b in hematopoietic cells and MLL fusion-mediated leukemia. MLL regulates expression of mir-196b and MLL fusion proteins cause a block in differentiation of progenitor cells. This effect may be due partially to high levels of mir-196b expression caused by the MLL fusion protein. High levels of mir-196b may also provide a survival advantage to cells expressing the MLL fusion protein.
Similar articles
- Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia.
Schotte D, Lange-Turenhout EA, Stumpel DJ, Stam RW, Buijs-Gladdines JG, Meijerink JP, Pieters R, Den Boer ML. Schotte D, et al. Haematologica. 2010 Oct;95(10):1675-82. doi: 10.3324/haematol.2010.023481. Epub 2010 May 21. Haematologica. 2010. PMID: 20494936 Free PMC article. - miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia.
Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, Jiang X, He C, Hyjek E, Zhang J, Zhang Z, Elkahloun A, Cao D, Shen C, Wunderlich M, Wang Y, Neilly MB, Jin J, Wei M, Lu J, Valk PJM, Delwel R, Lowenberg B, Le Beau MM, Vardiman J, Mulloy JC, Zeleznik-Le NJ, Liu PP, Zhang J, Chen J. Li Z, et al. Nat Commun. 2012 Feb 21;3:688. doi: 10.1038/ncomms1681. Nat Commun. 2012. PMID: 22353710 Free PMC article. - The PHD fingers of MLL block MLL fusion protein-mediated transformation.
Muntean AG, Giannola D, Udager AM, Hess JL. Muntean AG, et al. Blood. 2008 Dec 1;112(12):4690-3. doi: 10.1182/blood-2008-01-134056. Epub 2008 Sep 16. Blood. 2008. PMID: 18796627 Free PMC article. - Learning from mouse models of MLL fusion gene-driven acute leukemia.
Schwaller J. Schwaller J. Biochim Biophys Acta Gene Regul Mech. 2020 Aug;1863(8):194550. doi: 10.1016/j.bbagrm.2020.194550. Epub 2020 Apr 19. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32320749 Review. - Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins.
Ayton PM, Cleary ML. Ayton PM, et al. Oncogene. 2001 Sep 10;20(40):5695-707. doi: 10.1038/sj.onc.1204639. Oncogene. 2001. PMID: 11607819 Review.
Cited by
- microRNA expression in acute myeloid leukaemia: New targets for therapy?
Fletcher D, Brown E, Javadala J, Uysal-Onganer P, Guinn BA. Fletcher D, et al. EJHaem. 2022 Apr 26;3(3):596-608. doi: 10.1002/jha2.441. eCollection 2022 Aug. EJHaem. 2022. PMID: 36051053 Free PMC article. Review. - Transcriptional regulation of miR-196b by ETS2 in gastric cancer cells.
Liao YL, Hu LY, Tsai KW, Wu CW, Chan WC, Li SC, Lai CH, Ho MR, Fang WL, Huang KH, Lin WC. Liao YL, et al. Carcinogenesis. 2012 Apr;33(4):760-9. doi: 10.1093/carcin/bgs023. Epub 2012 Jan 31. Carcinogenesis. 2012. PMID: 22298639 Free PMC article. - The microRNA miR-196b acts as a tumor suppressor in Cdx2-driven acute myeloid leukemia.
Rawat VPS, Götze M, Rasalkar A, Vegi NM, Ihme S, Thoene S, Pastore A, Bararia D, Döhner H, Döhner K, Feuring-Buske M, Quintanilla-Fend L, Buske C. Rawat VPS, et al. Haematologica. 2020 Jun;105(6):e285-e289. doi: 10.3324/haematol.2019.223297. Epub 2019 Sep 26. Haematologica. 2020. PMID: 31558674 Free PMC article. No abstract available. - miRNA-mediated deregulation in leukemia.
Dell'aversana C, Altucci L. Dell'aversana C, et al. Front Genet. 2012 Nov 15;3:252. doi: 10.3389/fgene.2012.00252. eCollection 2012. Front Genet. 2012. PMID: 23372573 Free PMC article. - The stem cell-specific long noncoding RNA HOXA10-AS in the pathogenesis of KMT2A-rearranged leukemia.
Al-Kershi S, Bhayadia R, Ng M, Verboon L, Emmrich S, Gack L, Schwarzer A, Strowig T, Heckl D, Klusmann JH. Al-Kershi S, et al. Blood Adv. 2019 Dec 23;3(24):4252-4263. doi: 10.1182/bloodadvances.2019032029. Blood Adv. 2019. PMID: 31867596 Free PMC article.
References
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. - PubMed
- Rowley JD. The critical role of chromosome translocations in human leukemias. Annu Rev Genet. 1998;32:495–519. - PubMed
- Popovic R, Zeleznik L. MLL: how complex does it get? J Cell Biochem. 2005;95:234–242. - PubMed
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA105049/CA/NCI NIH HHS/United States
- CA105152/CA/NCI NIH HHS/United States
- R01 CA105152/CA/NCI NIH HHS/United States
- R01 HL087188/HL/NHLBI NIH HHS/United States
- P01 CA105049/CA/NCI NIH HHS/United States
- HL087188/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases