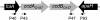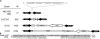The decay of the chromosomally encoded ccdO157 toxin-antitoxin system in the Escherichia coli species - PubMed (original) (raw)
The decay of the chromosomally encoded ccdO157 toxin-antitoxin system in the Escherichia coli species
Natacha Mine et al. Genetics. 2009 Apr.
Abstract
The origin and the evolution of toxin-antitoxin (TA) systems remain to be uncovered. TA systems are abundant in bacterial chromosomes and are thought to be part of the flexible genome that originates from horizontal gene transfer. To gain insight into TA system evolution, we analyzed the distribution of the chromosomally encoded ccdO157 system in 395 natural isolates of Escherichia coli. It was discovered in the E. coli O157:H7 strain in which it constitutes a genomic islet between two core genes (folA and apaH). Our study revealed that the folA-apaH intergenic region is plastic and subject to insertion of foreign DNA. It could be composed (i) of a repetitive extragenic palindromic (REP) sequence, (ii) of the ccdO157 system or subtle variants of it, (iii) of a large DNA piece that contained a ccdAO157 antitoxin remnant in association with ORFs of unknown function, or (iv) of a variant of it containing an insertion sequence in the ccdAO157 remnant. Sequence analysis and functional tests of the ccdO157 variants revealed that 69% of the variants were composed of an active toxin and antitoxin, 29% were composed of an active antitoxin and an inactive toxin, and in 2% of the cases both ORFs were inactive. Molecular evolution analysis showed that ccdBO157 is under neutral evolution, suggesting that this system is devoid of any biological role in the E. coli species.
Figures
Figure 1.—
PCR detection of the ccdO157 system in the E. coli species. The ccdO157 system is located between the folA and the apaH gene (not on scale). Its presence was detected by PCR using the P40 and P93 primers that are complementary to the ORFs that are flanking the ccdO157 system. Another set of primers (P43 and P47) complementary to the ccdAO157 and ccdBO157 ORFs was used to detect it at other potential chromosomal locations in strains that did not carry ccdO157 between folA and apaH.
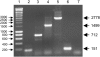
Figure 2.—
Diversity of the folA–apaH region in the E. coli species. PCR reactions were carried out on E. coli isolates using P40 and P93. Lane 1, molecular weight marker (SMART ladder, Eurogentec), size is indicated in base pairs; lane 2, MG1655; lane 3, O157 (2886-75, STEC Center); lane 4, O153 (LREC); lane 5, O163 (LREC); lane 6, O86 (LREC); lane 7, negative control. The size (base pairs) of the different amplicons is indicated.
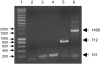
Figure 3.—
Diversity of the folA–apaH region within the O153 serogroup. PCR reactions were carried out on three O153 isolates using P40 and P93. Lane 1, molecular weight marker (SMART ladder, Eurogentec), size is indicated in base pairs; lane 2, negative control; lane 3, HC45; lane 4, MG1655; lane 5, HC71; lane 6, O153 (LREC). The size (base pairs) of the different amplicons is indicated.
Figure 4.—
Plasticity of the folA–apaH IR within the E. coli species. (A) Representation of the diversity of the folA–apaH IR. The strains and the size (base pairs) of the different folA–apaH intergenic regions (IR) are indicated. The folA and apaH genes are represented in black. The ccdAO157 and ccdBO157 genes are represented in white and dark gray, respectively (indicated as A and B). A′ represents the 3′-terminal part of the ccdAO157 gene. ORF2 (129 amino acids) is represented in light gray, ORF3 (199 amino acids) in white, and IS_621_ in stippled white. (B) Sequence alignment of the seq1 sequence and CcdAO157. The amino acid sequences were aligned with CLUSTALW. Symbols: asterisk, identical amino acids; colon, strongly similar amino acids; period, weakly similar amino acids. The total number of amino acids for each protein is in parentheses.
Figure 5.—
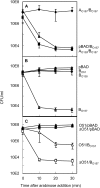
Properties of the ccdO51 and ccdO138 variants. (A) The CcdAO138 protein is not able to counteract the toxicity of CcdBO157. SG22622/pKK223-3/pBAD33-ccdBO157 (triangle), SG22622/pKK-ccdAO138/pBAD33-ccdBO157 (square), and SG22622/pKK-ccdAO157/pBAD33-ccdBO157 (diamond) were grown in the presence of 1% arabinose. (B) The CcdBO51 and CcdBO138 variants are not toxic. SG22622/pBAD33 (diamond), SG22622/pBAD33-ccdBO51 (square), SG22622/pBAD33-ccdBO138 (circle) and SG22622/pBAD33-ccdBO157 (triangle) were grown in the presence of 1% arabinose. In A and B serial dilutions of the cultures were plated at regular time intervals on CCM plates without arabinose and incubated overnight at 37°. Values correspond to the mean of results of three independent experiments and SDs are indicated. (C) The CcdAO51 protein is expressed in E. coli O51 and counteracts the toxicity of CcdBO157 expressed in trans. O51 (triangle) and O51Δ_ccdO51_ (square) containing the pBAD33 vector (solid symbols) or the pBAD33-ccdBO157 plasmid (open symbols) were grown in the presence of 0.25% arabinose. At regular time intervals, serial dilutions of the cultures were plated on CCM plates without arabinose and incubated overnight at 37°. Values correspond to the mean of results of three independent experiments and SDs are indicated.
Similar articles
- Functional interactions between coexisting toxin-antitoxin systems of the ccd family in Escherichia coli O157:H7.
Wilbaux M, Mine N, Guérout AM, Mazel D, Van Melderen L. Wilbaux M, et al. J Bacteriol. 2007 Apr;189(7):2712-9. doi: 10.1128/JB.01679-06. Epub 2007 Jan 26. J Bacteriol. 2007. PMID: 17259320 Free PMC article. - Activation of Toxin-Antitoxin System Toxins Suppresses Lethality Caused by the Loss of σE in Escherichia coli.
Daimon Y, Narita S, Akiyama Y. Daimon Y, et al. J Bacteriol. 2015 Jul;197(14):2316-24. doi: 10.1128/JB.00079-15. Epub 2015 Apr 27. J Bacteriol. 2015. PMID: 25917909 Free PMC article. - Genome-Wide Screening for Identification of Novel Toxin-Antitoxin Systems in Staphylococcus aureus.
Kato F, Yoshizumi S, Yamaguchi Y, Inouye M. Kato F, et al. Appl Environ Microbiol. 2019 Oct 1;85(20):e00915-19. doi: 10.1128/AEM.00915-19. Print 2019 Oct 15. Appl Environ Microbiol. 2019. PMID: 31375497 Free PMC article. - Bacterial toxin-antitoxin systems: more than selfish entities?
Van Melderen L, Saavedra De Bast M. Van Melderen L, et al. PLoS Genet. 2009 Mar;5(3):e1000437. doi: 10.1371/journal.pgen.1000437. Epub 2009 Mar 27. PLoS Genet. 2009. PMID: 19325885 Free PMC article. Review. - Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: hok/sok, ldr/rdl, symE/symR.
Kawano M. Kawano M. RNA Biol. 2012 Dec;9(12):1520-7. doi: 10.4161/rna.22757. Epub 2012 Nov 6. RNA Biol. 2012. PMID: 23131729 Review.
Cited by
- Toxin-antitoxin systems as multilevel interaction systems.
Goeders N, Van Melderen L. Goeders N, et al. Toxins (Basel). 2014 Jan 10;6(1):304-24. doi: 10.3390/toxins6010304. Toxins (Basel). 2014. PMID: 24434905 Free PMC article. Review. - Contribution of the Chromosomal ccdAB Operon to Bacterial Drug Tolerance.
Gupta K, Tripathi A, Sahu A, Varadarajan R. Gupta K, et al. J Bacteriol. 2017 Sep 5;199(19):e00397-17. doi: 10.1128/JB.00397-17. Print 2017 Oct 1. J Bacteriol. 2017. PMID: 28674066 Free PMC article. - Keeping the Wolves at Bay: Antitoxins of Prokaryotic Type II Toxin-Antitoxin Systems.
Chan WT, Espinosa M, Yeo CC. Chan WT, et al. Front Mol Biosci. 2016 Mar 22;3:9. doi: 10.3389/fmolb.2016.00009. eCollection 2016. Front Mol Biosci. 2016. PMID: 27047942 Free PMC article. Review. - Linkage, mobility, and selfishness in the MazF family of bacterial toxins: a snapshot of bacterial evolution.
Chopra N, Saumitra, Pathak A, Bhatnagar R, Bhatnagar S. Chopra N, et al. Genome Biol Evol. 2013;5(12):2268-84. doi: 10.1093/gbe/evt175. Genome Biol Evol. 2013. PMID: 24265503 Free PMC article. - Diversity, Prevalence, and Longitudinal Occurrence of Type II Toxin-Antitoxin Systems of Pseudomonas aeruginosa Infecting Cystic Fibrosis Lungs.
Andersen SB, Ghoul M, Griffin AS, Petersen B, Johansen HK, Molin S. Andersen SB, et al. Front Microbiol. 2017 Jun 23;8:1180. doi: 10.3389/fmicb.2017.01180. eCollection 2017. Front Microbiol. 2017. PMID: 28690609 Free PMC article.
References
- Bahassi, E. M., M. A. Salmon, L. Van Melderen, P. Bernard and M. Couturier, 1995. F plasmid CcdB killer protein: ccdB gene mutants coding for non-cytotoxic proteins which retain their regulatory functions. Mol. Microbiol. 15 1031–1037. - PubMed
- Bernard, P., and M. Couturier, 1991. The 41 carboxy-terminal residues of the miniF plasmid CcdA protein are sufficient to antagonize the killer activity of the CcdB protein. Mol. Gen. Genet. 226 297–304. - PubMed
- Christensen, S. K., K. Pedersen, F. G. Hansen and K. Gerdes, 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332 809–819. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources