The tree versus the forest: the fungal tree of life and the topological diversity within the yeast phylome - PubMed (original) (raw)
Comparative Study
The tree versus the forest: the fungal tree of life and the topological diversity within the yeast phylome
Marina Marcet-Houben et al. PLoS One. 2009.
Abstract
A recurrent topic in phylogenomics is the combination of various sequence alignments to reconstruct a tree that describes the evolutionary relationships within a group of species. However, such approach has been criticized for not being able to properly represent the topological diversity found among gene trees. To evaluate the representativeness of species trees based on concatenated alignments, we reconstruct several fungal species trees and compare them with the complete collection of phylogenies of genes encoded in the Saccharomyces cerevisiae genome. We found that, despite high levels of among-gene topological variation, the species trees do represent widely supported phylogenetic relationships. Most topological discrepancies between gene and species trees are concentrated in certain conflicting nodes. We propose to map such information on the species tree so that it accounts for the levels of congruence across the genome. We identified the lack of sufficient accuracy of current alignment and phylogenetic methods as an important source for the topological diversity encountered among gene trees. Finally, we discuss the implications of the high levels of topological variation for phylogeny-based orthology prediction strategies.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
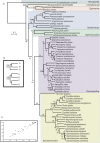
Figure 1. The Fungal species tree.
A) Phylogenetic tree representing the evolutionary relationships among the 60 fungal species considered in the study, as resulting from the ML analysis of the concatenated alignment of 69 widespread proteins. Numbers on the nodes indicate two different types of support values. The first number indicates the phylome support for that node, that is, the percentage of trees in the phylome that support the specific arrangement of the three or four groups of species defined by its daughter nodes (see B). An asterisk next to this number indicates that the topology obtained by the species tree is not the most common among the trees in the phylome. Whenever there is a second number (in bold), this indicates the bootstrap support when this is lower than 100. Partitions that do not have this second number have a bootstrap support of 100. Branches with dashed lines indicate evolutionary relationships that are supported by less than 50% of the trees in the phylome. B) Schematic representation of the two types of support values for the different nodes in the tree. X indicates the phylome support for the specific topology indicated by that node. Two types of nodes do exist attending to the number of partitions delimited by their daughter nodes. A first class of nodes (top), delimit relative topologies of three partitions (A, B and C), whereas a second class (bottom) delimit four partitions (A, B, C and D). Phylome support values indicate the percentage of trees that show exactly the relative grouping of the three or four groups delimited by the node. This percentage is expressed over the fraction of trees that contain at least one species from each of the partitions considered. The second number (Y) indicates the bootstrap support for the partition delimited by that node, but does not provide specific support for the specific arrangement of the sub-partitions within that partition. C) Correlation between the fungal species tree topologies recovered by the individual trees included in the concatenated alignment (Y axis) and all the trees in the phylome (X axis). In both cases the fraction of trees that are compatible with a given topology, as computed with the topology scanning algorithm, is represented.
Figure 2. Comparison of different orthology inference algorithms.
The synteny based and manually curated orthology predictions available at YGOB database is taken as a golden set to compute the number of true positives (TP), false positives (FP) and false negatives (FN) yielded by each method. For each method, the sensitivity S = TP/(TP+FN) and the positive predictive value P = TP/(TP+FP) are computed.
Similar articles
- PhylomeDB: a database for genome-wide collections of gene phylogenies.
Huerta-Cepas J, Bueno A, Dopazo J, Gabaldón T. Huerta-Cepas J, et al. Nucleic Acids Res. 2008 Jan;36(Database issue):D491-6. doi: 10.1093/nar/gkm899. Epub 2007 Oct 25. Nucleic Acids Res. 2008. PMID: 17962297 Free PMC article. - Comparative genomic hybridization provides new insights into the molecular taxonomy of the Saccharomyces sensu stricto complex.
Edwards-Ingram LC, Gent ME, Hoyle DC, Hayes A, Stateva LI, Oliver SG. Edwards-Ingram LC, et al. Genome Res. 2004 Jun;14(6):1043-51. doi: 10.1101/gr.2114704. Genome Res. 2004. PMID: 15173111 Free PMC article. - Assessment of phylogenomic and orthology approaches for phylogenetic inference.
Dutilh BE, van Noort V, van der Heijden RT, Boekhout T, Snel B, Huynen MA. Dutilh BE, et al. Bioinformatics. 2007 Apr 1;23(7):815-24. doi: 10.1093/bioinformatics/btm015. Epub 2007 Jan 19. Bioinformatics. 2007. PMID: 17237036 - The fascinating and secret wild life of the budding yeast S. cerevisiae.
Liti G. Liti G. Elife. 2015 Mar 25;4:e05835. doi: 10.7554/eLife.05835. Elife. 2015. PMID: 25807086 Free PMC article. Review. - Comparative genomics: a revolutionary tool for wine yeast strain development.
Borneman AR, Pretorius IS, Chambers PJ. Borneman AR, et al. Curr Opin Biotechnol. 2013 Apr;24(2):192-9. doi: 10.1016/j.copbio.2012.08.006. Epub 2012 Sep 1. Curr Opin Biotechnol. 2013. PMID: 22947601 Review.
Cited by
- Copper Acts Synergistically With Fluconazole in Candida glabrata by Compromising Drug Efflux, Sterol Metabolism, and Zinc Homeostasis.
Gaspar-Cordeiro A, Amaral C, Pobre V, Antunes W, Petronilho A, Paixão P, Matos AP, Pimentel C. Gaspar-Cordeiro A, et al. Front Microbiol. 2022 Jun 14;13:920574. doi: 10.3389/fmicb.2022.920574. eCollection 2022. Front Microbiol. 2022. PMID: 35774458 Free PMC article. - A phylogenomics approach for selecting robust sets of phylogenetic markers.
Capella-Gutierrez S, Kauff F, Gabaldón T. Capella-Gutierrez S, et al. Nucleic Acids Res. 2014 Apr;42(7):e54. doi: 10.1093/nar/gku071. Epub 2014 Jan 29. Nucleic Acids Res. 2014. PMID: 24476915 Free PMC article. - Systems biology of host-fungus interactions: turning complexity into simplicity.
Tierney L, Kuchler K, Rizzetto L, Cavalieri D. Tierney L, et al. Curr Opin Microbiol. 2012 Aug;15(4):440-6. doi: 10.1016/j.mib.2012.05.001. Epub 2012 Jun 19. Curr Opin Microbiol. 2012. PMID: 22717554 Free PMC article. Review. - Complete DNA sequence of Kuraishia capsulata illustrates novel genomic features among budding yeasts (Saccharomycotina).
Morales L, Noel B, Porcel B, Marcet-Houben M, Hullo MF, Sacerdot C, Tekaia F, Leh-Louis V, Despons L, Khanna V, Aury JM, Barbe V, Couloux A, Labadie K, Pelletier E, Souciet JL, Boekhout T, Gabaldon T, Wincker P, Dujon B. Morales L, et al. Genome Biol Evol. 2013;5(12):2524-39. doi: 10.1093/gbe/evt201. Genome Biol Evol. 2013. PMID: 24317973 Free PMC article. - Finding single copy genes out of sequenced genomes for multilocus phylogenetics in non-model fungi.
Feau N, Decourcelle T, Husson C, Desprez-Loustau ML, Dutech C. Feau N, et al. PLoS One. 2011 Apr 13;6(4):e18803. doi: 10.1371/journal.pone.0018803. PLoS One. 2011. PMID: 21533204 Free PMC article.
References
- Eisen JA. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. - PubMed
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases