Accelerating molecular dynamic simulation on graphics processing units - PubMed (original) (raw)
Accelerating molecular dynamic simulation on graphics processing units
Mark S Friedrichs et al. J Comput Chem. 2009.
Abstract
We describe a complete implementation of all-atom protein molecular dynamics running entirely on a graphics processing unit (GPU), including all standard force field terms, integration, constraints, and implicit solvent. We discuss the design of our algorithms and important optimizations needed to fully take advantage of a GPU. We evaluate its performance, and show that it can be more than 700 times faster than a conventional implementation running on a single CPU core.
(c) 2009 Wiley Periodicals, Inc.
Figures
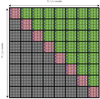
Figure 1
The set of pxp tiles required for force calculation-only the pink and green tiles need to be calculated. Force data for the grey tiles can be generated by negating the sign of the forces calculated for the corresponding green tile on the other side of the pink diagonal.
Similar articles
- Accelerating the Generalized Born with Molecular Volume and Solvent Accessible Surface Area Implicit Solvent Model Using Graphics Processing Units.
Gong X, Chiricotto M, Liu X, Nordquist E, Feig M, Brooks CL 3rd, Chen J. Gong X, et al. J Comput Chem. 2020 Mar 30;41(8):830-838. doi: 10.1002/jcc.26133. Epub 2019 Dec 24. J Comput Chem. 2020. PMID: 31875339 Free PMC article. - CUSA and CUDE: GPU-accelerated methods for estimating solvent accessible surface area and desolvation.
Dynerman D, Butzlaff E, Mitchell JC. Dynerman D, et al. J Comput Biol. 2009 Apr;16(4):523-37. doi: 10.1089/cmb.2008.0157. J Comput Biol. 2009. PMID: 19361325 - Inference of dynamic spatial GRN models with multi-GPU evolutionary computation.
Mousavi R, Konuru SH, Lobo D. Mousavi R, et al. Brief Bioinform. 2021 Sep 2;22(5):bbab104. doi: 10.1093/bib/bbab104. Brief Bioinform. 2021. PMID: 33834216 - Graphics processing units in bioinformatics, computational biology and systems biology.
Nobile MS, Cazzaniga P, Tangherloni A, Besozzi D. Nobile MS, et al. Brief Bioinform. 2017 Sep 1;18(5):870-885. doi: 10.1093/bib/bbw058. Brief Bioinform. 2017. PMID: 27402792 Free PMC article. Review. - GPU computing for systems biology.
Dematté L, Prandi D. Dematté L, et al. Brief Bioinform. 2010 May;11(3):323-33. doi: 10.1093/bib/bbq006. Epub 2010 Mar 7. Brief Bioinform. 2010. PMID: 20211843 Review.
Cited by
- Sintering Rate and Mechanism of TiO2 Nanoparticles by Molecular Dynamics.
Buesser B, Gröhn AJ, Pratsinis SE. Buesser B, et al. J Phys Chem C Nanomater Interfaces. 2011 Jun 9;115(22):11030-11035. doi: 10.1021/jp2032302. J Phys Chem C Nanomater Interfaces. 2011. PMID: 23730400 Free PMC article. - The native GCN4 leucine-zipper domain does not uniquely specify a dimeric oligomerization state.
Oshaben KM, Salari R, McCaslin DR, Chong LT, Horne WS. Oshaben KM, et al. Biochemistry. 2012 Nov 27;51(47):9581-91. doi: 10.1021/bi301132k. Epub 2012 Nov 13. Biochemistry. 2012. PMID: 23116373 Free PMC article. - Integrating atomistic molecular dynamics simulations, experiments, and network analysis to study protein dynamics: strength in unity.
Papaleo E. Papaleo E. Front Mol Biosci. 2015 May 27;2:28. doi: 10.3389/fmolb.2015.00028. eCollection 2015. Front Mol Biosci. 2015. PMID: 26075210 Free PMC article. - Efficient nonbonded interactions for molecular dynamics on a graphics processing unit.
Eastman P, Pande VS. Eastman P, et al. J Comput Chem. 2010 Apr 30;31(6):1268-72. doi: 10.1002/jcc.21413. J Comput Chem. 2010. PMID: 19847780 Free PMC article. - GPU-Accelerated Molecular Dynamics and Free Energy Methods in Amber18: Performance Enhancements and New Features.
Lee TS, Cerutti DS, Mermelstein D, Lin C, LeGrand S, Giese TJ, Roitberg A, Case DA, Walker RC, York DM. Lee TS, et al. J Chem Inf Model. 2018 Oct 22;58(10):2043-2050. doi: 10.1021/acs.jcim.8b00462. Epub 2018 Sep 25. J Chem Inf Model. 2018. PMID: 30199633 Free PMC article.
References
- Elsen E, Houston M, Vishal V, Darve E, Hanrahan P, Pande VS. SC06 Proceedings. 2006
- Stone JE, Phillips JC, Freddolino PL, Hardy DJ, Trabuco LG, Schulten K. J Comput Chem. 2007;28:2618. - PubMed
- Owens JD, Luebke D, Govindaraju N, Harris M, Krüger J, Lefohn AE, Purcell T. J Comput Graphics Forum. 2007;26:80.
- Hess B, Kutzner C, van der Spoel D, Lindahl E. J Chem Theory Comput. 2008;4:435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM062868/GM/NIGMS NIH HHS/United States
- R01 GM062868-07/GM/NIGMS NIH HHS/United States
- U54 GM072970/GM/NIGMS NIH HHS/United States
- R01-GM062868/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources