Alternative pre-mRNA splicing switches modulate gene expression in late erythropoiesis - PubMed (original) (raw)
Alternative pre-mRNA splicing switches modulate gene expression in late erythropoiesis
Miki L Yamamoto et al. Blood. 2009.
Abstract
Differentiating erythroid cells execute a unique gene expression program that insures synthesis of the appropriate proteome at each stage of maturation. Standard expression microarrays provide important insight into erythroid gene expression but cannot detect qualitative changes in transcript structure, mediated by RNA processing, that alter structure and function of encoded proteins. We analyzed stage-specific changes in the late erythroid transcriptome via use of high-resolution microarrays that detect altered expression of individual exons. Ten differentiation-associated changes in erythroblast splicing patterns were identified, including the previously known activation of protein 4.1R exon 16 splicing. Six new alternative splicing switches involving enhanced inclusion of internal cassette exons were discovered, as well as 3 changes in use of alternative first exons. All of these erythroid stage-specific splicing events represent activated inclusion of authentic annotated exons, suggesting they represent an active regulatory process rather than a general loss of splicing fidelity. The observation that 3 of the regulated transcripts encode RNA binding proteins (SNRP70, HNRPLL, MBNL2) may indicate significant changes in the RNA processing machinery of late erythroblasts. Together, these results support the existence of a regulated alternative pre-mRNA splicing program that is critical for late erythroid differentiation.
Figures
Figure 1
Stage-specific switch of protein 4.1R exon 16 splicing in highly purified erythroblast cultures. (A) Fluorescence-activated cell sorter analysis of day 7 erythroblasts from 3 different preparations indicates that purity is more than or equal to 97%, based on expression of erythroid markers for glycophorin A and CD71. Quantitative analysis demonstrated erythroblast purities as follows: prep 1, 97%; prep 2, 97%; prep 3, 99%. (B) RT-PCR scheme used to analyze 4.1R pre-mRNA splicing in early (day 7) and late erythroblasts (day 14), using primers in the nearest constitutive exons 13 and 17. Gel image shows primarily exclusion of exon 16 in early erythroblasts (bottom band), whereas substantial inclusion of exon 16 was observed in late erythroblasts (top band). Alternative exons 14 and 15 are not expressed in erythroid cells.
Figure 2
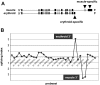
Exon array detection of erythroid-specific beta spectrin mRNA 3′ end. (A) The exon structure of human beta spectrin transcripts in muscle versus erythroid cells, which express distinct 3′ terminal exons. (B) The splicing index of probe sets across the full length of the beta spectrin gene. Numbers along the horizontal axis represent probe set IDs. Positive values for the splicing index represent higher relative probe set expression in erythroblasts, whereas negative values indicate higher relative expression in muscle. The significant upward peak maps to the known erythroid-specific 3′ end, whereas the downward peak represents the muscle-specific 3′ end.
Figure 3
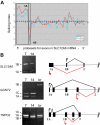
Stage-specific change in expression of alternative first exons. (A) Splicing index shows normalized changes in probe set expression along the entire SLC12A6 transcript for day 10 (blue curve) and day 14 erythroblasts (red curve), relative to expression at day 7 (black line). Results show a stage-dependent increase in expression of probe sets representing the 1A region (boxed) and a decrease in expression of probe sets for exon 1B. (B) RT-PCR validation of exon array predictions for SLC12A6 and 2 additional genes. Shown are gels of PCR products validating alternative splicing switches in first exon expression (left) and diagrams of the relevant pre-mRNA regions (right). Black arrow/black lines represent predominant pattern in day 7 erythroblasts; red arrow/red lines, predominant pattern in day 14 erythroblasts. Alternative first exons are indicated by 1A, 1B, 1C, and a shared constitutive exon indicated as exon 2. Common names of the alternatively spliced transcripts are as follows: SLC12A6 indicates KCl cotransporter 3 (KCC3); TNPO2, transportin 2 (a nuclear import protein); GCNT2, glucosaminyl (N-acetyl) transferase 2 (generates the branched chain carbohydrate structure that constitutes the I antigen).
Figure 4
Exon junction array detection of the 4.1R exon 16 splicing switch. Probe sets in the exon 16 region that exhibit significant changes in relative expression between day 7 and day 14 were mapped to the human genome using the UCSC BLAT alignment tool. (Bottom) The exon structure of the 2 mRNA isoforms expressed from the 4.1R gene; arrow represents alternative exon 16. (Top) Probe sets interrogating the exon 16 inclusion event were up-regulated at day 14, whereas a reciprocal decrease in expression of the exons 13 to 17 skipping event was observed. RT-PCR validation of this splicing switch is shown in Figure 1. Alternative exons 14 and 15 are not expressed in these cultures.
Figure 5
Novel stage-specific alternative splicing switches in erythroid genes. (A) General scheme for detection of alternative splicing of a pre-mRNA (left) into the mRNA isoform including the alternative exon (top right) or the mRNA skipping this exon (bottom right). Diagnostic isoform-specific microarray probes are indicated above the spliced mRNAs, whereas PCR primers used for validation are shown below the mRNAs (arrows). In addition, there are exon probes for the first and third exons that hybridize equally to both isoforms and are useful for determining overall transcript levels. (B) Shown are gels of PCR products validating alternative splicing switches in late erythropoiesis (left) and diagrams of the relevant pre-mRNA regions (right). Gels demonstrate substantial increases in exon inclusion products (top bands, indicated by arrows), relative to exon-skipping products (bottom bands), at day 14. The deduced splicing patterns are indicated at the right, with black lines indicating major splice patterns at day 7 and red lines indicating predominant splice pattern at day 14. Asterisks indicate positions of stop codons (not shown for ARFIP1 and PLD1 because they are located farther downstream). Common names of the alternatively spliced transcripts are as follows: HNRPLL indicates heterogeneous nuclear ribonucleoprotein L-like (an hnRNP protein); SNRP70, U1 small nuclear ribonucleoprotein 70K (a component of the U1 snRNP); MBNL2, muscleblind 2 (RNA binding proteins with known splicing regulatory activity); ATP11C, ATPase class VI type 11C; ARFIP1, ADP-ribosylation factor interacting protein 1; PLD1, phospholipase D1.
Similar articles
- A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis.
Pimentel H, Parra M, Gee S, Ghanem D, An X, Li J, Mohandas N, Pachter L, Conboy JG. Pimentel H, et al. Nucleic Acids Res. 2014 Apr;42(6):4031-42. doi: 10.1093/nar/gkt1388. Epub 2014 Jan 17. Nucleic Acids Res. 2014. PMID: 24442673 Free PMC article. - The role of alternative pre-mRNA splicing in regulating the structure and function of skeletal protein 4.1.
Conboy J. Conboy J. Proc Soc Exp Biol Med. 1999 Feb;220(2):73-8. doi: 10.1046/j.1525-1373.1999.d01-12.x. Proc Soc Exp Biol Med. 1999. PMID: 10049099 Review. - Decrease in hnRNP A/B expression during erythropoiesis mediates a pre-mRNA splicing switch.
Hou VC, Lersch R, Gee SL, Ponthier JL, Lo AJ, Wu M, Turck CW, Koury M, Krainer AR, Mayeda A, Conboy JG. Hou VC, et al. EMBO J. 2002 Nov 15;21(22):6195-204. doi: 10.1093/emboj/cdf625. EMBO J. 2002. PMID: 12426391 Free PMC article. - RNA splicing during terminal erythropoiesis.
Conboy JG. Conboy JG. Curr Opin Hematol. 2017 May;24(3):215-221. doi: 10.1097/MOH.0000000000000329. Curr Opin Hematol. 2017. PMID: 28118223 Free PMC article. Review. - Regulation of alternative pre-mRNA splicing during erythroid differentiation.
Hou VC, Conboy JG. Hou VC, et al. Curr Opin Hematol. 2001 Mar;8(2):74-9. doi: 10.1097/00062752-200103000-00003. Curr Opin Hematol. 2001. PMID: 11224680 Review.
Cited by
- Efficient in vivo manipulation of alternative pre-mRNA splicing events using antisense morpholinos in mice.
Parra MK, Gee S, Mohandas N, Conboy JG. Parra MK, et al. J Biol Chem. 2011 Feb 25;286(8):6033-9. doi: 10.1074/jbc.M110.158154. Epub 2010 Dec 14. J Biol Chem. 2011. PMID: 21156798 Free PMC article. - Spliceosomal component Sf3b1 is essential for hematopoietic differentiation in zebrafish.
De La Garza A, Cameron RC, Nik S, Payne SG, Bowman TV. De La Garza A, et al. Exp Hematol. 2016 Sep;44(9):826-837.e4. doi: 10.1016/j.exphem.2016.05.012. Epub 2016 Jun 1. Exp Hematol. 2016. PMID: 27260753 Free PMC article. - Is there a direct role for erythrocytes in the immune response?
Morera D, MacKenzie SA. Morera D, et al. Vet Res. 2011 Jul 29;42(1):89. doi: 10.1186/1297-9716-42-89. Vet Res. 2011. PMID: 21801407 Free PMC article. Review. - Evolution of alternative splicing in primate brain transcriptomes.
Lin L, Shen S, Jiang P, Sato S, Davidson BL, Xing Y. Lin L, et al. Hum Mol Genet. 2010 Aug 1;19(15):2958-73. doi: 10.1093/hmg/ddq201. Epub 2010 May 11. Hum Mol Genet. 2010. PMID: 20460271 Free PMC article. - The cardiotonic steroid digitoxin regulates alternative splicing through depletion of the splicing factors SRSF3 and TRA2B.
Anderson ES, Lin CH, Xiao X, Stoilov P, Burge CB, Black DL. Anderson ES, et al. RNA. 2012 May;18(5):1041-9. doi: 10.1261/rna.032912.112. Epub 2012 Mar 28. RNA. 2012. PMID: 22456266 Free PMC article.
References
- Johnson JM, Castle J, Garrett-Engele P, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. - PubMed
- Hou VC, Conboy JG. Regulation of alternative pre-mRNA splicing during erythroid differentiation. Curr Opin Hematol. 2001;8:74–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 DK075021/DK/NIDDK NIH HHS/United States
- R56 HL045182/HL/NHLBI NIH HHS/United States
- DK75021/DK/NIDDK NIH HHS/United States
- R01 HL045182/HL/NHLBI NIH HHS/United States
- HL45182/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous