Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation - PubMed (original) (raw)
. 2009 Feb 5;360(6):599-605.
doi: 10.1056/NEJMoa0805392.
Julie Gauthier, Dan Spiegelman, Anne Noreau, Yan Yang, Stéphanie Pellerin, Sylvia Dobrzeniecka, Mélanie Côté, Elizabeth Perreau-Linck, Lionel Carmant, Guy D'Anjou, Eric Fombonne, Anjene M Addington, Judith L Rapoport, Lynn E Delisi, Marie-Odile Krebs, Faycal Mouaffak, Ridha Joober, Laurent Mottron, Pierre Drapeau, Claude Marineau, Ronald G Lafrenière, Jean Claude Lacaille, Guy A Rouleau, Jacques L Michaud; Synapse to Disease Group
Affiliations
- PMID: 19196676
- PMCID: PMC2925262
- DOI: 10.1056/NEJMoa0805392
Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation
Fadi F Hamdan et al. N Engl J Med. 2009.
Erratum in
- N Engl J Med. 2009 Oct 29;361(18):1814. Perreault-Linck, Elizabeth [corrected to Perreau-Linck, Elizabeth]
Abstract
Although autosomal forms of nonsyndromic mental retardation account for the majority of cases of mental retardation, the genes that are involved remain largely unknown. We sequenced the autosomal gene SYNGAP1, which encodes a ras GTPase-activating protein that is critical for cognition and synapse function, in 94 patients with nonsyndromic mental retardation. We identified de novo truncating mutations (K138X, R579X, and L813RfsX22) in three of these patients. In contrast, we observed no de novo or truncating mutations in SYNGAP1 in samples from 142 subjects with autism spectrum disorders, 143 subjects with schizophrenia, and 190 control subjects. These results indicate that SYNGAP1 disruption is a cause of autosomal dominant nonsyndromic mental retardation.
2009 Massachusetts Medical Society
Conflict of interest statement
No potential conflict of interest relevant to this article was reported.
Figures
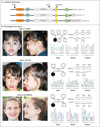
Figure 1. De Novo SYNGAP1 Mutations in Three Patients with Nonsyndromic Mental Retardation
Panel A shows the localization of de novo SYNGAP1 mutations identified in patients with nonsyndromic mental retardation. Amino acid positions are based on the NCBI Reference Sequence project number NP_006763 (from NM_006772) (for isoform 1, which consists of 1343 amino acids). The various predicted functional domains are highlighted: pleckstrin homology domain (PH; positions 150 to 251), C2 domain (positions 263 to 362), RASGAP (positions 392 to 729), SH3 (positions 785 to 815), CC domain (positions 1189 to 1262), T/SXV type 1 PDZ-binding motif (QTRV) (isoform 2), and CamKII binding (GAAPGPPRHG) (isoform 3). In addition, the last amino acids of isoform 1 (TADH) are shown. The variable C-terminals of the three SYNGAP1 isoforms that are shown here correspond to GenBank complementary DNA accession numbers AB067525 (for isoform 1), AK307888 (for isoform 2), and AL713634 (for isoform 3). The hatched lines for isoforms 2 and 3 indicate that the amino acid sequence has been abbreviated. Panel B shows chromatograms corresponding to the SYNGAP1 sequence for each of the three patients with de novo mutations and for their parents. Wild-type and mutant SYNGAP1 DNA sequences are shown, along with the corresponding amino acids.
Comment in
- SYNGAP: bridging the gap between genetic factors and autosomal non-syndromic mental retardation.
Huang K. Huang K. Clin Genet. 2009 Aug;76(2):149-51. doi: 10.1111/j.1399-0004.2009.01247_3.x. Epub 2009 Aug 7. Clin Genet. 2009. PMID: 19673947 No abstract available.
Similar articles
- De novo SYNGAP1 mutations in nonsyndromic intellectual disability and autism.
Hamdan FF, Daoud H, Piton A, Gauthier J, Dobrzeniecka S, Krebs MO, Joober R, Lacaille JC, Nadeau A, Milunsky JM, Wang Z, Carmant L, Mottron L, Beauchamp MH, Rouleau GA, Michaud JL. Hamdan FF, et al. Biol Psychiatry. 2011 May 1;69(9):898-901. doi: 10.1016/j.biopsych.2010.11.015. Epub 2011 Jan 15. Biol Psychiatry. 2011. PMID: 21237447 - Mutations in SYNGAP1 cause intellectual disability, autism, and a specific form of epilepsy by inducing haploinsufficiency.
Berryer MH, Hamdan FF, Klitten LL, Møller RS, Carmant L, Schwartzentruber J, Patry L, Dobrzeniecka S, Rochefort D, Neugnot-Cerioli M, Lacaille JC, Niu Z, Eng CM, Yang Y, Palardy S, Belhumeur C, Rouleau GA, Tommerup N, Immken L, Beauchamp MH, Patel GS, Majewski J, Tarnopolsky MA, Scheffzek K, Hjalgrim H, Michaud JL, Di Cristo G. Berryer MH, et al. Hum Mutat. 2013 Feb;34(2):385-94. doi: 10.1002/humu.22248. Epub 2012 Dec 12. Hum Mutat. 2013. PMID: 23161826 - [Autosomal dominant mental retardation type 5 caused by SYNGAP1 gene mutations: a report of 8 cases and literature review].
Wang XL, Tian YN, Chen C, Peng J. Wang XL, et al. Zhongguo Dang Dai Er Ke Za Zhi. 2023 May 15;25(5):489-496. doi: 10.7499/j.issn.1008-8830.2301054. Zhongguo Dang Dai Er Ke Za Zhi. 2023. PMID: 37272175 Free PMC article. Review. Chinese. - Novel Mutation of SYNGAP1 Associated with Autosomal Dominant Mental Retardation 5 in a Chinese Patient.
Pei Y, Li W, Du L, Wei F. Pei Y, et al. Fetal Pediatr Pathol. 2018 Dec;37(6):400-403. doi: 10.1080/15513815.2018.1497113. Epub 2018 Dec 21. Fetal Pediatr Pathol. 2018. PMID: 30572772 - SYNGAP1 mutations: Clinical, genetic, and pathophysiological features.
Agarwal M, Johnston MV, Stafstrom CE. Agarwal M, et al. Int J Dev Neurosci. 2019 Nov;78:65-76. doi: 10.1016/j.ijdevneu.2019.08.003. Epub 2019 Aug 24. Int J Dev Neurosci. 2019. PMID: 31454529 Review.
Cited by
- Transition between synaptic branch formation and synaptogenesis is regulated by the lin-4 microRNA.
Xu Y, Quinn CC. Xu Y, et al. Dev Biol. 2016 Dec 1;420(1):60-66. doi: 10.1016/j.ydbio.2016.10.010. Epub 2016 Oct 13. Dev Biol. 2016. PMID: 27746167 Free PMC article. - SynGAP isoforms differentially regulate synaptic plasticity and dendritic development.
Araki Y, Hong I, Gamache TR, Ju S, Collado-Torres L, Shin JH, Huganir RL. Araki Y, et al. Elife. 2020 Jun 24;9:e56273. doi: 10.7554/eLife.56273. Elife. 2020. PMID: 32579114 Free PMC article. - SYNGAP1 links the maturation rate of excitatory synapses to the duration of critical-period synaptic plasticity.
Clement JP, Ozkan ED, Aceti M, Miller CA, Rumbaugh G. Clement JP, et al. J Neurosci. 2013 Jun 19;33(25):10447-52. doi: 10.1523/JNEUROSCI.0765-13.2013. J Neurosci. 2013. PMID: 23785156 Free PMC article. - The role of altered translation in intellectual disability and epilepsy.
Malone TJ, Kaczmarek LK. Malone TJ, et al. Prog Neurobiol. 2022 Jun;213:102267. doi: 10.1016/j.pneurobio.2022.102267. Epub 2022 Mar 29. Prog Neurobiol. 2022. PMID: 35364140 Free PMC article. Review. - A Selective Review of the Excitatory-Inhibitory Imbalance in Schizophrenia: Underlying Biology, Genetics, Microcircuits, and Symptoms.
Liu Y, Ouyang P, Zheng Y, Mi L, Zhao J, Ning Y, Guo W. Liu Y, et al. Front Cell Dev Biol. 2021 Oct 21;9:664535. doi: 10.3389/fcell.2021.664535. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34746116 Free PMC article. Review.
References
- Chelly J, Khelfaoui M, Francis F, Chérif B, Bienvenu T. Genetics and pathophysiology of mental retardation. Eur J Hum Genet. 2006;14:701–713. - PubMed
- Ropers HH, Hamel BC. X-linked mental retardation. Nat Rev Genet. 2005;6:46–57. - PubMed
- Basel-Vanagaite L. Genetics of autosomal recessive non-syndromic mental retardation: recent advances. Clin Genet. 2007;72:167–174. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases