Mutant presenilin 1 increases the expression and activity of BACE1 - PubMed (original) (raw)
. 2009 Apr 3;284(14):9027-38.
doi: 10.1074/jbc.M805685200. Epub 2009 Feb 5.
Roberta Borghi, Alessandra Piccini, Rosa Mangerini, Sandro Sorbi, Gabriella Cirmena, Anna Garuti, Bernardino Ghetti, Fabrizio Tagliavini, Mohamed R Mughal, Mark P Mattson, Xiongwei Zhu, Xinglong Wang, Michela Guglielmotto, Elena Tamagno, Massimo Tabaton
Affiliations
- PMID: 19196715
- PMCID: PMC2666551
- DOI: 10.1074/jbc.M805685200
Mutant presenilin 1 increases the expression and activity of BACE1
Luca Giliberto et al. J Biol Chem. 2009.
Abstract
Mutations of the presenilin 1 (PS1) gene are the most common cause of early onset familial Alzheimer disease (FAD). PS1 mutations alter the activity of the gamma-secretase on the beta-amyloid precursor protein (APP), leading to selective overproduction of beta-amyloid (Abeta) 42 peptides, the species that forms oligomers that may exert toxic effects on neurons. Here we show that PS1 mutations, expressed both transiently and stably, in non-neuronal and neuronal cell lines increase the expression and the activity of the beta-secretase (BACE1), the rate-limiting step of Abeta production. Also, BACE1 expression and activity are elevated in brains of PS1 mutant knock-in mice compared with wild type littermates as well as in cerebral cortex of FAD cases bearing various PS1 mutations compared with in sporadic AD cases and controls. The up-regulation of BACE1 by PS1 mutations requires the gamma-secretase cleavage of APP and is proportional to the amount of secreted Abeta42. Abeta42, and not AICD (APP intracellular domain), is indeed the APP derivative that mediates the overexpression of BACE1. The effect of PS1 mutations on BACE1 may contribute to determine the wide clinical and pathological phenotype of early onset FAD.
Figures
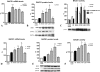
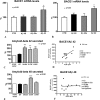
FIGURE 1.
PS1 mutations induce the transcription and activity of BACE1. A–C, HEK-293 APPwt cells were transfected with empty vector, wild type PS1, or mutant PS1 constructs. BACE1 mRNA expression (A), BACE1 protein levels (B) and activity (C) were analyzed; also, indication of BACE1 activity is given by the analysis of βAPPs on cell culture media. D–F, MEFs deficient for PS1 were transfected with control, wild type, and mutant plasmids. Quantification of BACE1 mRNA (D), protein levels (E), and activity (F) was performed. Data represent the mean and S.E. of three or more experiments; significance is relative to empty vector control. Statistical analysis was performed with ANOVA and Bonferroni post-test.
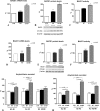
FIGURE 2.
PS1 mutations stably expressed in neuronal cell lines determine BACE1 up-regulation. SH-SY5Y, stably transfected with pcDNA3.1, PS1 wt, and PS1 mutant S170F, were analyzed for BACE1 mRNA (A), BACE1 protein levels (B), and BACE1 activity (C). Similarly, M17 neuronal cells, stably expressing PS1 A246E mutant, PS1 wt, and pcDNA3.1, were analyzed: D, BACE1 mRNA; E, BACE1 protein levels; F, BACE1 activity.G and H, sandwich enzyme-linked immunosorbent assay for Aβ40 and -42, performed analyzing conditioned medium of SH-SY5Y stable cells (G) and M17 stable cells (F). The Aβ42/40 ratio is also presented. Data represent the mean and S.E. of three or more experiments, performed on two different stable cell clones; significance is relative to empty vector control (CTR); for enzyme-linked immunosorbent assay, p values are specified in the figure. Statistical analysis was performed with ANOVA and Bonferroni post-test.
FIGURE 3.
BACE1 expression and activity are significantly increased in PS1 mutant knock-in mice and in PS1 mutant FAD cases. A and B, M146V knock in (PS1 M146V KI) mice (4 males and 4 females) and age-matched littermates (4 males and 3 females) were analyzed. mRNA was extracted from one dissected brain hemisphere, whereas protein extraction was performed on the other hemisphere. A, BACE1 mRNA. B, BACE1 protein levels.C, an aliquot of brain protein lysates was also used to perform BACE1 activity assay. D and E, frozen brains from 11 FAD patients carrying 10 different mutations, 10 sporadic AD (SAD) patients, and 12 control (CTR) subjects were processed likewise. D, BACE1 mRNA levels; E, BACE1 activity. Statistical analysis was performed with t test (A–C) and ANOVA (D_–_E) and the Bonferroni post-test.
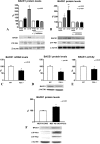
FIGURE 4.
The up-regulation of BACE1 induced by PS1 mutations requires the activity of the γ-secretase. A, HEK-293 APPwt cells were treated with a transition analogue inhibitor of γ-secretase at the moment of transfection with empty vector control, PS1 wild type, and PS1 S170F and replenished twice every 4 h; BACE1 protein levels were analyzed.B, MEFs PS1–_/– were treated similarly, and BACE1 protein levels were analyzed. Basal levels of BACE1 mRNA (C), protein levels (D), and activity (E) were evaluated in MEFs PS1–/–, all resulting lower than in wild type cells. F, MEFs deficient for both PS1 and PS2 were transfected with empty vector control or with PS1 and PS2: reconstitution of PS1/2 expression determined an increased expression of BACE1; PS1/2 CTFs are produced as expected. Data represent the mean and S.E. of three or more experiments; significance is relative to the empty vector control or to control cells. Statistical analysis was performed with ANOVA (A and_B) and Bonferroni post-test and t test (C–F).
FIGURE 5.
The activation of BACE1 by mutant PS1 requires APP. MEFs from APP/APLP2-deficient mice were transfected with empty vector control, wild type, and all the mutants considered in Fig. 1. Furthermore, MEFs APP DKO were transfected with human APP 695, reconstituting its expression, and with a representative PS1 mutant (S170F). Quantification of BACE1 mRNA (A), protein levels (B), and activity (C) were carried out as previously described. D–F, APP knock down (E, Western blot shown, performed with APP N-terminal-specific antibody) was performed on HEK-293 cells. Cells were transfected with non-targeting siRNA or Homo sapien APP-specific siRNA in 2 pulses, at times 0 and 24 h later; empty vector control, PS1 wild type, and PS1 S170F mutant were transfected subsequently, and quantification of BACE1 mRNA (D), protein levels (E), and activity (F) were performed. Data represent the mean and S.E. of three or more experiments; significance is relative to empty vector control. Statistical analysis was performed with ANOVA and the Bonferroni post-test. CTR, control.
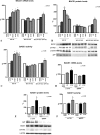
FIGURE 6.
APP intracellular derivatives do not play a role in BACE1 transcriptional control. A, a series of neuronal and non neuronal cell lines (SKNBE(2c), SH-SY5Y, HEK-293, and MEFs) was transiently transfected with AICD derived from γ-or ε-secretase cleavage of APP. BACE1 transcription levels were determined. Data are a pool of several experimental repeats, performed at least in three repeats for each different cell line, which yielded analogous results. Transfected AICD transcript was detected by PCR conducted on the same cDNAs used for BACE1 real time PCR experiments, and a representative panel of semiquantitative PCR is shown. B, wild type and Fe65-deficient MEFs were transfected with empty vector, PS1 wt, or PS1 S170F. BACE1 transcription levels were determined. Data represent the mean and S.E. of three or more experiments; significance is relative to empty vector control. C, seven 4-week-old AICD transgenic mice (tg) and their littermates were analyzed for BACE1 mRAN expression. AICD length (50, 57, or 59 amino acids), mouse sex (F or M) and tg line (1-5, 5-1, 4-4) are indicated. Data from tg mice are compared with each corresponding littermate control (each control (CTR) = 100). Statistical analysis was performed with ANOVA and Bonferroni post-test.
FIGURE 7.
Aβ-(1–42) peptide is the APP derivative responsible for the up-regulation of BACE1. A, primary neurons, obtained from a 17.5 pregnant FVB mouse, and cultured for 21 days (similar data for 14 days in vitro neurons not shown) were treated for 1 h with freshly dissolved 1 μ
m
Aβ peptides or with medium alone. BACE1 transcription levels were determined. Nine wells for each condition from three different neuronal cultures were analyzed as single experimental points. B, a series of neuronal cell lines (SKNBE(2c) and SH-SY5Y) were treated similarly at least 18 h after plating with 1 μ
m
Aβ peptides or with medium alone for 1 h. mRNA was extracted as described, and TaqMan Real Time PCR for BACE1 was performed. Data are a pool of several experimental repeats, performed at least in three repeats for each different cell line, which yielded analogous results; they represent the mean and S.E. C and_E_, HEK-293 APPwt cells were transfected with empty vector, wild type PS1, and mutant PS1 constructs. Aβ species were quantified in conditioned media by enzyme-linked immunosorbent assay. D and F, BACE1 activity was also measured in these cells and correlated to the levels of Aβ40 and Aβ42. Statistical analysis was performed with ANOVA and the Bonferroni post-test and the study of linear correlation (D and_F_). CTR, control.
Similar articles
- PS1 Affects the Pathology of Alzheimer's Disease by Regulating BACE1 Distribution in the ER and BACE1 Maturation in the Golgi Apparatus.
Li N, Qiu Y, Wang H, Zhao J, Qing H. Li N, et al. Int J Mol Sci. 2022 Dec 18;23(24):16151. doi: 10.3390/ijms232416151. Int J Mol Sci. 2022. PMID: 36555791 Free PMC article. - NFkappaB-dependent control of BACE1 promoter transactivation by Abeta42.
Buggia-Prevot V, Sevalle J, Rossner S, Checler F. Buggia-Prevot V, et al. J Biol Chem. 2008 Apr 11;283(15):10037-47. doi: 10.1074/jbc.M706579200. Epub 2008 Feb 8. J Biol Chem. 2008. PMID: 18263584 - Calpain activation promotes BACE1 expression, amyloid precursor protein processing, and amyloid plaque formation in a transgenic mouse model of Alzheimer disease.
Liang B, Duan BY, Zhou XP, Gong JX, Luo ZG. Liang B, et al. J Biol Chem. 2010 Sep 3;285(36):27737-44. doi: 10.1074/jbc.M110.117960. Epub 2010 Jul 1. J Biol Chem. 2010. PMID: 20595388 Free PMC article. - BACE1: the beta-secretase enzyme in Alzheimer's disease.
Vassar R. Vassar R. J Mol Neurosci. 2004;23(1-2):105-14. doi: 10.1385/JMN:23:1-2:105. J Mol Neurosci. 2004. PMID: 15126696 Review. - The beta-secretase, BACE: a prime drug target for Alzheimer's disease.
Vassar R. Vassar R. J Mol Neurosci. 2001 Oct;17(2):157-70. doi: 10.1385/JMN:17:2:157. J Mol Neurosci. 2001. PMID: 11816789 Review.
Cited by
- Amyloid-β production: major link between oxidative stress and BACE1.
Tamagno E, Guglielmotto M, Monteleone D, Tabaton M. Tamagno E, et al. Neurotox Res. 2012 Oct;22(3):208-19. doi: 10.1007/s12640-011-9283-6. Epub 2011 Oct 15. Neurotox Res. 2012. PMID: 22002808 Review. - Molecular basis of etiological implications in Alzheimer's disease: focus on neuroinflammation.
Niranjan R. Niranjan R. Mol Neurobiol. 2013 Dec;48(3):412-28. doi: 10.1007/s12035-013-8428-4. Epub 2013 Feb 19. Mol Neurobiol. 2013. PMID: 23420079 Review. - Signaling effect of amyloid-beta(42) on the processing of AbetaPP.
Tabaton M, Zhu X, Perry G, Smith MA, Giliberto L. Tabaton M, et al. Exp Neurol. 2010 Jan;221(1):18-25. doi: 10.1016/j.expneurol.2009.09.002. Epub 2009 Sep 9. Exp Neurol. 2010. PMID: 19747481 Free PMC article. Review. - Store-Operated Calcium Entry Inhibition and Plasma Membrane Calcium Pump Upregulation Contribute to the Maintenance of Resting Cytosolic Calcium Concentration in A1-like Astrocytes.
Poejo J, Berrocal M, Saez L, Gutierrez-Merino C, Mata AM. Poejo J, et al. Molecules. 2023 Jul 12;28(14):5363. doi: 10.3390/molecules28145363. Molecules. 2023. PMID: 37513235 Free PMC article. - Distinct patterns of APP processing in the CNS in autosomal-dominant and sporadic Alzheimer disease.
Pera M, Alcolea D, Sánchez-Valle R, Guardia-Laguarta C, Colom-Cadena M, Badiola N, Suárez-Calvet M, Lladó A, Barrera-Ocampo AA, Sepulveda-Falla D, Blesa R, Molinuevo JL, Clarimón J, Ferrer I, Gelpi E, Lleó A. Pera M, et al. Acta Neuropathol. 2013 Feb;125(2):201-13. doi: 10.1007/s00401-012-1062-9. Epub 2012 Dec 6. Acta Neuropathol. 2013. PMID: 23224319 Free PMC article.
References
- Haass, C., Schlossmacher, M. G., Hung, A. Y., Vigo-Pelfrey, C., Mellon, A., Ostaszewski, B. L., Lieberburg, I., Koo, E. H., Schenk, D., Teplow, D. B., and Selkoe, D. J. (1992) Nature 359 322–325 - PubMed
- Wolfe, M. S., Xia, W., Ostaszewski, B. L., Diehl, T. S., Kimberly, W. T., and Selkoe, D. J. (1999) Nature 398 513–517 - PubMed
- Scheuner, D., Eckman, C., Jensen, M., Song, X., Citron, M., Suzuki, N., Bird, T. D., Hardy, J., Hutton, M., Kukull, W., Larson, E., Levy-Lahad, E., Viitanen, M., Peskind, E., Poorkaj, P., Schellenberg, G., Tanzi, R., Wasco, W., Lannfelt, L., Selkoe, D., and Younkin, S. (1996) Nat. Med. 2 864–870 - PubMed
- Xia, W., Zhang, J., Kholodenko, D., Citron, M., Podlisny, M. B., Teplow, D. B., Haass, C., Seubert, P., Koo, E. H., and Selkoe, D. J. (1997) J. Biol. Chem. 272 7977–7982 - PubMed
- Teller, J. K., Russo, C., DeBusk, L. M., Angelini, G., Zaccheo, D., Dagna-Bricarelli, F., Scartezzini, P., Bertolini, S., Mann, D. M., Tabaton, M., and Gambetti, P. (1996) Nat. Med. 2 93–95 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources