Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin - PubMed (original) (raw)
Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin
Laura C Wieland Brown et al. Proc Natl Acad Sci U S A. 2009.
Abstract
The thiazolylpeptides are a family of >50 bactericidal antibiotics that block the initial steps of bacterial protein synthesis. Here, we report a biosynthetic gene cluster for thiocillin and establish that it, and by extension the whole class, is ribosomally synthesized. Remarkably, the C-terminal 14 residues of a 52-residue peptide precursor undergo 13 posttranslational modifications to give rise to thiocillin, making this antibiotic the most heavily posttranslationally-modified peptide known to date.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Schematic of the thiocillin gene cluster. A 22-kb gene cluster from B. cereus ATCC 14579 encodes 24 genes responsible for the production of the thiocillins. Four identical structural genes encode a 52-residue peptide, of which the last 14 residues undergo 13 posttranslational modifications of 6 varieties to become the mature thiocillins. Gray squares indicate the R groups in the thiocillin family members. The peptide residues are numbered starting with the first residue after the leader peptide; thus, Ser-39 = Ser-1.
Fig. 2.
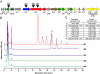
Characterization of thiocillins by HPLC and MS. (A) Schematic of the thiocillin gene cluster, showing the plasmid integration sites of insertional mutants IM1, IM2, and IM4, and control strain IM3. (B) HPLC stack plot showing methanolic extracts of cell material from cultures of B. cereus ATCC 14579, insertional mutants IM1, IM2, IM4, and control strain IM3. Peaks corresponding to compounds 1–8 are labeled. Thiocillin production is abolished in the insertional mutants, whereas in IM3, flux is shifted toward the nonhydroxylated thiocillins 1, 2, 5, and 8. (C) High-resolution MS data for compounds 1–8.
Fig. 3.
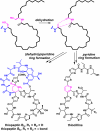
Cryptic dehydration in the proposed mechanism for pyridine and dehydropiperidine ring formation. Dehydration of Ser-1 and Ser-10 yields Dha residues at both positions. Subsequent cyclization leads to cleavage of the leader peptide and formation of the fully aromatic pyridine in the thiocillins, whereas a portion of the leader peptide is retained and the nonaromatic piperidine or dehydropiperidine is formed in the thiopeptins. The pyridine and (dehydro)piperidine rings are shown in pink, and the leader peptide fragment and leader peptide/quinaldic acid loop are shown in blue.
Fig. 4.
Thiazolylpeptide and related gene clusters. The gene clusters for thiocillin and goadsporin and the predicted gene cluster for berninamycin are shown at the top; the structural peptide sequence is shown below the strain name, with the small molecule-encoding C-terminal sequence highlighted in red. Ten other gene clusters harboring homologs of both the heterocycle-forming (tclJ/tclN) and the Dha/Dhb-forming (tclK/tclL) genes are shown.
Similar articles
- Generation of thiocillin variants by prepeptide gene replacement and in vivo processing by Bacillus cereus.
Acker MG, Bowers AA, Walsh CT. Acker MG, et al. J Am Chem Soc. 2009 Dec 9;131(48):17563-5. doi: 10.1021/ja908777t. J Am Chem Soc. 2009. PMID: 19911780 Free PMC article. - Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications.
Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. Liao R, et al. Chem Biol. 2009 Feb 27;16(2):141-7. doi: 10.1016/j.chembiol.2009.01.007. Chem Biol. 2009. PMID: 19246004 Free PMC article. - Manipulation of thiocillin variants by prepeptide gene replacement: structure, conformation, and activity of heterocycle substitution mutants.
Bowers AA, Acker MG, Koglin A, Walsh CT. Bowers AA, et al. J Am Chem Soc. 2010 Jun 2;132(21):7519-27. doi: 10.1021/ja102339q. J Am Chem Soc. 2010. PMID: 20455532 Free PMC article. - Thiazolyl peptide antibiotic biosynthesis: a cascade of post-translational modifications on ribosomal nascent proteins.
Walsh CT, Acker MG, Bowers AA. Walsh CT, et al. J Biol Chem. 2010 Sep 3;285(36):27525-31. doi: 10.1074/jbc.R110.135970. Epub 2010 Jun 3. J Biol Chem. 2010. PMID: 20522549 Free PMC article. Review. - Discovery of novel fungal RiPP biosynthetic pathways and their application for the development of peptide therapeutics.
Vogt E, Künzler M. Vogt E, et al. Appl Microbiol Biotechnol. 2019 Jul;103(14):5567-5581. doi: 10.1007/s00253-019-09893-x. Epub 2019 May 31. Appl Microbiol Biotechnol. 2019. PMID: 31147756 Review.
Cited by
- The posttranslational modification cascade to the thiopeptide berninamycin generates linear forms and altered macrocyclic scaffolds.
Malcolmson SJ, Young TS, Ruby JG, Skewes-Cox P, Walsh CT. Malcolmson SJ, et al. Proc Natl Acad Sci U S A. 2013 May 21;110(21):8483-8. doi: 10.1073/pnas.1307111110. Epub 2013 May 6. Proc Natl Acad Sci U S A. 2013. PMID: 23650400 Free PMC article. - Saturation mutagenesis of TsrA Ala4 unveils a highly mutable residue of thiostrepton A.
Zhang F, Kelly WL. Zhang F, et al. ACS Chem Biol. 2015 Apr 17;10(4):998-1009. doi: 10.1021/cb5007745. Epub 2015 Jan 20. ACS Chem Biol. 2015. PMID: 25572285 Free PMC article. - Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining.
Skinnider MA, Johnston CW, Edgar RE, Dejong CA, Merwin NJ, Rees PN, Magarvey NA. Skinnider MA, et al. Proc Natl Acad Sci U S A. 2016 Oct 18;113(42):E6343-E6351. doi: 10.1073/pnas.1609014113. Epub 2016 Oct 3. Proc Natl Acad Sci U S A. 2016. PMID: 27698135 Free PMC article. - Isolation and characterization of the gene cluster for biosynthesis of the thiopeptide antibiotic TP-1161.
Engelhardt K, Degnes KF, Zotchev SB. Engelhardt K, et al. Appl Environ Microbiol. 2010 Nov;76(21):7093-101. doi: 10.1128/AEM.01442-10. Epub 2010 Sep 17. Appl Environ Microbiol. 2010. PMID: 20851988 Free PMC article. - Identification and analysis of the biosynthetic gene cluster encoding the thiopeptide antibiotic cyclothiazomycin in Streptomyces hygroscopicus 10-22.
Wang J, Yu Y, Tang K, Liu W, He X, Huang X, Deng Z. Wang J, et al. Appl Environ Microbiol. 2010 Apr;76(7):2335-44. doi: 10.1128/AEM.01790-09. Epub 2010 Feb 12. Appl Environ Microbiol. 2010. PMID: 20154110 Free PMC article.
References
- Baldwin JE, Abraham E. The biosynthesis of penicillins and cephalosporins. Natural Product Rep. 1988;5:129–145. - PubMed
- Donadio S, Sosio M, Stegmann E, Weber T, Wohlleben W. Comparative analysis and insights into the evolution of gene clusters for glycopeptide antibiotic biosynthesis. Mol Genet Genomics. 2005;274:40–50. - PubMed
- Baltz RH. Biosynthesis and genetic engineering of lipopeptide antibiotics related to daptomycin. Curr Top Med Chem. 2008;8:618–638. - PubMed
- Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides 1. Annu Rev Microbiol. 2004;58:453–488. - PubMed
- Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM020011/GM/NIGMS NIH HHS/United States
- R01 CA059021/CA/NCI NIH HHS/United States
- R01 GM049338/GM/NIGMS NIH HHS/United States
- GM49338/GM/NIGMS NIH HHS/United States
- R01 GM086258/GM/NIGMS NIH HHS/United States
- GM20011/GM/NIGMS NIH HHS/United States
- R01 CA024487/CA/NCI NIH HHS/United States
- CA59021/CA/NCI NIH HHS/United States
- R01 GM086258-02/GM/NIGMS NIH HHS/United States
- CA24487/CA/NCI NIH HHS/United States
- R01 GM020011/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources