An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans - PubMed (original) (raw)
An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans
Rui Lu et al. PLoS Pathog. 2009 Feb.
Abstract
Dicer ribonucleases of plants and invertebrate animals including Caenorhabditis elegans recognize and process a viral RNA trigger into virus-derived small interfering RNAs (siRNAs) to guide specific viral immunity by Argonaute-dependent RNA interference (RNAi). C. elegans also encodes three Dicer-related helicase (drh) genes closely related to the RIG-I-like RNA helicase receptors which initiate broad-spectrum innate immunity against RNA viruses in mammals. Here we developed a transgenic C. elegans strain that expressed intense green fluorescence from a chromosomally integrated flock house virus replicon only after knockdown or knockout of a gene required for antiviral RNAi. Use of the reporter nematode strain in a feeding RNAi screen identified drh-1 as an essential component of the antiviral RNAi pathway. However, RNAi induced by either exogenous dsRNA or the viral replicon was enhanced in drh-2 mutant nematodes, whereas exogenous RNAi was essentially unaltered in drh-1 mutant nematodes, indicating that exogenous and antiviral RNAi pathways are genetically distinct. Genetic epistatic analysis shows that drh-1 acts downstream of virus sensing and viral siRNA biogenesis to mediate specific antiviral RNAi. Notably, we found that two members of the substantially expanded subfamily of Argonautes specific to C. elegans control parallel antiviral RNAi pathways. These findings demonstrate both conserved and unique strategies of C. elegans in antiviral defense.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
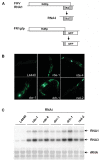
Figure 1. Screening for viral immunity genes in C. elegans by feeding RNAi.
(A) Genome structure and expression of wildtype FHV RNA1 (FR1) and its B2-deficient mutant, FR1gfp, that expresses the enhanced GFP in place of B2. (B) Detection of green fluorescence in FR1gfp reporter worms after feeding RNAi targeting specific genes or the commonly used L4440 vector as indicated, photographed 48 hours after induction of the replicon replication. (C) Accumulation of FR1gfp genomic (RNA1) and subgenomic RNA (RNA3) by northern blotting in FR1gf worms with (lanes 3–12) and without (lanes 1–2) feeding RNAi of specific worm genes. Two independent tests were analyzed for each E. coli strain. Methylene blue staining of total RNA was provided to show equal loading.
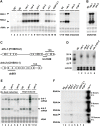
Figure 2. Molecular and functional characterization of the drh-1 and drh-2 genes.
(A) Accumulation of the FR1gfp replicon RNAs in genetic mutant worm strains carrying the same FR1gfp transgene array. Total RNA was also analyzed from wildtype N2 worms with (N2) and without the FR1gfp transgene (N2*) 48 hours after induction of the replicon replication (h.a.i.). (B) Induction of the RNAi immunity by the replicon in a worm integrant different from that analyzed in (A). (C) Molecular structures and genetic lesions of the drh-1 and drh-2 genes. (D) Accumulation of (-) viral siRNAs in single knockout worm mutants 48 hours after induction of the replicon replication. 45 µg of total small RNAs was loaded in each lane. A combination of 18 32P end-labeled DNA oligos corresponding to eGFP coding sequence in tandem was used as the probe for viral siRNA detection. The same filters were probed for miR-58 after stripping as the loading control. (E) Detection of drh-1 and drh-2 transcripts before and after induction of the replicon replication. Two independent tests were analyzed for each strain. (F) Time course analysis of the accumulation of the replicon RNAs in wildtype and mutant worms 2, 6, and 16 h.a.i. Total RNA extracted from FR1gfp rde-1 worms 48 h.a.i. was loaded as a control (lane rde-1*). Methylene blue staining of total RNA was provided to show equal loading.
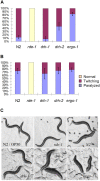
Figure 3. drh-2 is a negative regulator of exogenous RNAi.
(A and B) unc-22 RNAi phenotype in response to unc-22 dsRNA microinjected at 25 and 100 µg/ml, respectively. 30 to 40 worms were used for unc-22 dsRNA injection. Shown here are the percentages of twitching and paralyzed F1 progenies of each injected worm collected between 8 and 32 hours post injection. The error bars indicate standard deviation for the paralysis phenotype. (C) Morphological phenotype of the F1 progenies of wildtype and mutant worms after feeding RNAi targeting dpy-13. All worm strains were synchronized before feeding RNAi.
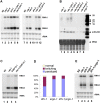
Figure 4. Analysis of endogenous small RNAs.
(A) Accumulation of K02E2.6 and X-cluster siRNAs as well as miR-58, miR-238, and let-7 and lin-4 miRNAs in wildtype and mutant worm strains with (lane 1) or without the FR1gfp transgene (lanes 2–9). 45 µg of total small RNAs was loaded in each lane. The same filter was repeatedly reprobed after stripping. Small RNAs loaded in lane 1 was extracted from FR1gfp worms 48 hours post induction. (B) Accumulation of 21U1 piRNA in wildtype and mutant worm strains in the absence of FR1gfp transgene. End-labeled DNA oligos complementary to endo-siRNAs, miRNA and 21U1 piRNA were used as the probes. Ethidium bromide staining of tRNAs was provided to show equal loading.
Figure 5. Genetic epistatic analysis of antiviral RNAi.
(A, C, E) Accumulation of the FR1gfp replicon RNAs in single and double knockout worm mutants 48 after replicon replication. (B) Accumulation of (-) viRNAs in single and double knockout worm mutants 48 after replicon replication as described in Figure 2. A combination of 18 32P end-labeled DNA oligos corresponding to eGFP coding sequence in tandem was used as the probe for viral siRNA detection. The same filters were probed for miR-238 after stripping. (D) unc-22 RNAi phenotype in response to unc-22 dsRNA microinjected at 100 µg/ml as described in Figure 3.
Similar articles
- Caenorhabditis elegans RIG-I Homolog Mediates Antiviral RNA Interference Downstream of Dicer-Dependent Biogenesis of Viral Small Interfering RNAs.
Coffman SR, Lu J, Guo X, Zhong J, Jiang H, Broitman-Maduro G, Li WX, Lu R, Maduro M, Ding SW. Coffman SR, et al. mBio. 2017 Mar 21;8(2):e00264-17. doi: 10.1128/mBio.00264-17. mBio. 2017. PMID: 28325765 Free PMC article. - Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms.
Guo X, Zhang R, Wang J, Ding SW, Lu R. Guo X, et al. Proc Natl Acad Sci U S A. 2013 Oct 1;110(40):16085-90. doi: 10.1073/pnas.1307453110. Epub 2013 Sep 16. Proc Natl Acad Sci U S A. 2013. PMID: 24043766 Free PMC article. - Transgene-Assisted Genetic Screen Identifies rsd-6 and Novel Genes as Key Components of Antiviral RNA Interference in Caenorhabditis elegans.
Long T, Meng F, Lu R. Long T, et al. J Virol. 2018 Aug 16;92(17):e00416-18. doi: 10.1128/JVI.00416-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29950414 Free PMC article. - Shared and unique mechanisms of RNAi-mediated antiviral immunity in C. elegans.
Yan T, Lu R. Yan T, et al. Virology. 2025 Apr;605:110459. doi: 10.1016/j.virol.2025.110459. Epub 2025 Feb 21. Virology. 2025. PMID: 40022946 Review. - Biology and Mechanisms of Short RNAs in Caenorhabditis elegans.
Grishok A. Grishok A. Adv Genet. 2013;83:1-69. doi: 10.1016/B978-0-12-407675-4.00001-8. Adv Genet. 2013. PMID: 23890211 Review.
Cited by
- Caenorhabditis elegans as a model for intracellular pathogen infection.
Balla KM, Troemel ER. Balla KM, et al. Cell Microbiol. 2013 Aug;15(8):1313-22. doi: 10.1111/cmi.12152. Epub 2013 May 13. Cell Microbiol. 2013. PMID: 23617769 Free PMC article. Review. - RNA interference may result in unexpected phenotypes in Caenorhabditis elegans.
De-Souza EA, Camara H, Salgueiro WG, Moro RP, Knittel TL, Tonon G, Pinto S, Pinca APF, Antebi A, Pasquinelli AE, Massirer KB, Mori MA. De-Souza EA, et al. Nucleic Acids Res. 2019 May 7;47(8):3957-3969. doi: 10.1093/nar/gkz154. Nucleic Acids Res. 2019. PMID: 30838421 Free PMC article. - Engineering recombinant Orsay virus directly in the metazoan host Caenorhabditis elegans.
Jiang H, Franz CJ, Wang D. Jiang H, et al. J Virol. 2014 Oct;88(20):11774-81. doi: 10.1128/JVI.01630-14. Epub 2014 Jul 30. J Virol. 2014. PMID: 25078701 Free PMC article. - Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans.
Rechavi O, Minevich G, Hobert O. Rechavi O, et al. Cell. 2011 Dec 9;147(6):1248-56. doi: 10.1016/j.cell.2011.10.042. Epub 2011 Nov 23. Cell. 2011. PMID: 22119442 Free PMC article. - A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity.
Ashe A, Bélicard T, Le Pen J, Sarkies P, Frézal L, Lehrbach NJ, Félix MA, Miska EA. Ashe A, et al. Elife. 2013 Oct 8;2:e00994. doi: 10.7554/eLife.00994. Elife. 2013. PMID: 24137537 Free PMC article.
References
- Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. - PubMed
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. - PubMed
- Yoneyama M, Fujita T. Structural Mechanism of RNA Recognition by the RIG-I-like Receptors. Immunity. 2008;29:178–181. - PubMed
- Hammond SM. Dicing and Slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials