Poly(dA:dT) tracts: major determinants of nucleosome organization - PubMed (original) (raw)
Review
Poly(dA:dT) tracts: major determinants of nucleosome organization
Eran Segal et al. Curr Opin Struct Biol. 2009 Feb.
Abstract
Homopolymeric stretches of deoxyadenosine nucleotides (A's) on one strand of double-stranded DNA, referred to as poly(dA:dT) tracts or A-tracts, are overabundant in eukaryotic genomes. They have unusual structural, dynamic, and mechanical properties, and may resist sharp bending. Such unusual material properties, together with their overabundance in eukaryotes, raised the possibility that poly(dA:dT) tracts might function in eukaryotes to influence the organization of nucleosomes at many genomic regions. Recent genome-wide studies strongly confirm these ideas and suggest that these tracts play major roles in chromatin organization and genome function. Here we review what is known about poly(dA:dT) tracts and how they work.
Figures
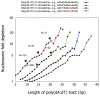
Figure 1. Nucleosome are relatively depleted over poly(dA:dT) tracts in vivo
Shown is the combined nucleosome fold depletion over all poly(dA:dT) tracts of length k, for k = 5,6,7,…, and for tracts with exactly 0, 2, 4, or 6 base substitutions. Each graph is trimmed at a length K at which there are less than 10 such tracts in the S. cerevisiae genome, and the fold depletion at this final point is computed over all elements whose length is at least K. The number of underlying elements at various points in the graph is indicated (N). Figure adapted from ref. [**9].
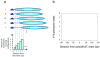
Figure 2. Poly(dA:dT) tracts create larger nucleosome-depleted regions
(a) Shown is a simple example focusing only on the immediate neighborhood of the boundary. All (five) possible nucleosome configurations are illustrated, in which a nucleosome (cyan ovals) can be placed within five basepairs of the boundary (blue triangle). The number and set of nucleosome configurations occupying each of the five basepairs immediately adjacent to the boundary are shown in the graph below. If all configurations are equally likely, then basepairs closer to the poly(dA:dT) tract will exhibit lower nucleosome occupancy simply because fewer nucleosome configurations cover those basepairs [22]. (b) Schematic showing that nucleosome depletion caused by a poly(dA:dT) tract is maximal over the tract itself, but extends for considerable distances in either direction. Thus specific factor DNA binding sites located nearby to a poly(dA:dT) tract will have relatively enhanced accessibility compared to factor sites located far from a poly(dA:dT) tract, facilitating binding of the factor. Panel (a) adapted from ref. [**9].
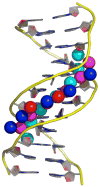
Figure 3. Narrow minor groove and multilayer spine of hydration in a poly(dA:dT) tract
Shown is a representation of the atomic resolution X-ray crystallographic structure of [d(CGCAAATTTGCG)]2 [**44]. The DNA backbones are shown as yellow curves, with the bases shown in a partial-charge-coded stick representation. The narrow minor groove of the A3T3 stretch has many high occupancy water molecules, 4 layers deep, shown here as spheres, color coded according to their layer from innermost to outermost as cyan, purple, blue, and red, respectively. The multiple layers, extensive hydrogen bonding, and high occupancy of these waters all suggest that they may have strongly favorable energetic interactions with themselves and the DNA. Figure kindly provided by Prof. L.D. Williams (Georgia Tech.).
Similar articles
- Destabilization of nucleosomes by an unusual DNA conformation adopted by poly(dA) small middle dotpoly(dT) tracts in vivo.
Shimizu M, Mori T, Sakurai T, Shindo H. Shimizu M, et al. EMBO J. 2000 Jul 3;19(13):3358-65. doi: 10.1093/emboj/19.13.3358. EMBO J. 2000. PMID: 10880448 Free PMC article. - Positioned and G/C-capped poly(dA:dT) tracts associate with the centers of nucleosome-free regions in yeast promoters.
Wu R, Li H. Wu R, et al. Genome Res. 2010 Apr;20(4):473-84. doi: 10.1101/gr.103226.109. Epub 2010 Feb 4. Genome Res. 2010. PMID: 20133331 Free PMC article. - Modulation of cyclobutane pyrimidine dimer formation in a positioned nucleosome containing poly(dA.dT) tracts.
Schieferstein U, Thoma F. Schieferstein U, et al. Biochemistry. 1996 Jun 18;35(24):7705-14. doi: 10.1021/bi953011r. Biochemistry. 1996. PMID: 8672471 - The Active Mechanism of Nucleosome Depletion by Poly(dA:dT) Tracts In Vivo.
Barnes T, Korber P. Barnes T, et al. Int J Mol Sci. 2021 Jul 30;22(15):8233. doi: 10.3390/ijms22158233. Int J Mol Sci. 2021. PMID: 34360997 Free PMC article. Review. - A structural perspective on the where, how, why, and what of nucleosome positioning.
Arya G, Maitra A, Grigoryev SA. Arya G, et al. J Biomol Struct Dyn. 2010 Jun;27(6):803-20. doi: 10.1080/07391102.2010.10508585. J Biomol Struct Dyn. 2010. PMID: 20232935 Review.
Cited by
- A computational approach to map nucleosome positions and alternative chromatin states with base pair resolution.
Zhou X, Blocker AW, Airoldi EM, O'Shea EK. Zhou X, et al. Elife. 2016 Sep 13;5:e16970. doi: 10.7554/eLife.16970. Elife. 2016. PMID: 27623011 Free PMC article. - Distal chromatin structure influences local nucleosome positions and gene expression.
Jansen A, van der Zande E, Meert W, Fink GR, Verstrepen KJ. Jansen A, et al. Nucleic Acids Res. 2012 May;40(9):3870-85. doi: 10.1093/nar/gkr1311. Epub 2012 Jan 12. Nucleic Acids Res. 2012. PMID: 22241769 Free PMC article. - Probing a label-free local bend in DNA by single molecule tethered particle motion.
Brunet A, Chevalier S, Destainville N, Manghi M, Rousseau P, Salhi M, Salomé L, Tardin C. Brunet A, et al. Nucleic Acids Res. 2015 Jun 23;43(11):e72. doi: 10.1093/nar/gkv201. Epub 2015 Mar 12. Nucleic Acids Res. 2015. PMID: 25765645 Free PMC article. - Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast.
Raveh-Sadka T, Levo M, Shabi U, Shany B, Keren L, Lotan-Pompan M, Zeevi D, Sharon E, Weinberger A, Segal E. Raveh-Sadka T, et al. Nat Genet. 2012 May 27;44(7):743-50. doi: 10.1038/ng.2305. Nat Genet. 2012. PMID: 22634752 - Open chromatin structures regulate the efficiencies of pre-RC formation and replication initiation in Epstein-Barr virus.
Papior P, Arteaga-Salas JM, Günther T, Grundhoff A, Schepers A. Papior P, et al. J Cell Biol. 2012 Aug 20;198(4):509-28. doi: 10.1083/jcb.201109105. Epub 2012 Aug 13. J Cell Biol. 2012. PMID: 22891264 Free PMC article.
References
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci USA. 1985;82:8419–8423. This paper was the first to report that poly(dA:dT) elements in a gene’s promoter facilitate transcription in vivo, and the first to connect this observation with a possible mechanism, suggesting that poly(dA:dT) elements could act by depleting repressive nucleosomes from the promoter. - PMC - PubMed
- Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. This paper was the first to provide concrete evidence in support of the model in which poly(dA:dT) elements act by depleting repressive nucleosomes from a gene’s promoter, thereby effectively enhancing transcription. - PMC - PubMed
- Moreira JM, Remacle JE, Kielland-Brandt MC, Holmberg S. Datin, a yeast poly(dA:dT)-binding protein, behaves as an activator of the wild-type ILV1 promoter and interacts synergistically with Reb1p. Mol Gen Genet. 1998;258:95–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM054692/GM/NIGMS NIH HHS/United States
- R01 GM054692-11/GM/NIGMS NIH HHS/United States
- R01 GM058617/GM/NIGMS NIH HHS/United States
- R01 GM058617-11/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources