The extent of population genetic subdivision differs among four co-distributed shark species in the Indo-Australian archipelago - PubMed (original) (raw)
The extent of population genetic subdivision differs among four co-distributed shark species in the Indo-Australian archipelago
Jenny R Ovenden et al. BMC Evol Biol. 2009.
Abstract
Background: The territorial fishing zones of Australia and Indonesia are contiguous to the north of Australia in the Timor and Arafura Seas and in the Indian Ocean to the north of Christmas Island. The area surrounding the shared boundary consists of a variety of bio-diverse marine habitats including shallow continental shelf waters, oceanic trenches and numerous offshore islands. Both countries exploit a variety of fisheries species, including whaler (Carcharhinus spp.) and hammerhead sharks (Sphyrna spp.). Despite their differences in social and financial arrangements, the two countries are motivated to develop complementary co-management practices to achieve resource sustainability. An essential starting point is knowledge of the degree of population subdivision, and hence fisheries stock status, in exploited species.
Results: Populations of four commercially harvested shark species (Carcharhinus obscurus, Carcharhinus sorrah, Prionace glauca, Sphyrna lewini) were sampled from northern Australia and central Indonesia. Neutral genetic markers (mitochondrial DNA control region sequence and allelic variation at co-dominant microsatellite loci) revealed genetic subdivision between Australian and Indonesian populations of C. sorrah. Further research is needed to address the possibility of genetic subdivision among C. obscurus populations. There was no evidence of genetic subdivision for P. glauca and S. lewini populations, but the sampling represented a relatively small part of their distributional range. For these species, more detailed analyses of population genetic structure is recommended in the future.
Conclusion: Cooperative management between Australia and Indonesia is the best option at present for P. glauca and S. lewini, while C. sorrah and C. obscurus should be managed independently. On-going research on these and other exploited shark and ray species is strongly recommended. Biological and ecological similarity between species may not be a predictor of population genetic structure, so species-specific studies are recommended to provide new data to assist with sustainable fisheries management.
Figures
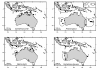
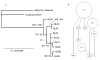
Figure 1
Circles encompass collection locations for four shark species from East Australia (1), West Australia (2), Indonesia (3), Gulf of Carpentaria (4) and mid-north Pacific (5).
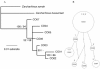
Figure 2
Inferred phylogeny (A) and statistical parsimony network (B) among haplotypes of C. obscurus. The phylogeny was rooted with C. sorrah and C. dussumieri and nodal support is given as Bayesian posterior probabilities/ML boostrap support. Dash (-) indicates support of less than 50%. In the network, each indicated step (circle) represents a single nucleotide difference in the mtDNA control region sequence. The area of circles is scaled to represent the relative frequency of that haplotype and the smallest circle represent inferred haplotypes that were not sampled. The collection location of sampled haplotypes is numbered (in italics) according to Fig. 1.
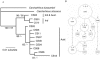
Figure 3
Inferred phylogeny (A) and statistical parsimony network (B) among haplotypes of C. sorrah collected in Indonesia (Ind) and Australia (Aust). The phylogeny was rooted with C. obscurus and C. dussumieri and nodal support is given as Bayesian posterior probabilities/ML boostrap support. Dash (-) indicates support of less than 50%. In the network, each indicated step (circle) represents a single nucleotide difference in the mtDNA control region sequence. The area of circles is scaled to represent the relative frequency of that haplotype and the smallest circle represent inferred haplotypes that were not sampled. The collection location of sampled haplotypes is numbered (in italics) according to Fig. 1.
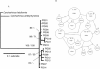
Figure 4
Inferred phylogeny (A) and statistical parsimony network (B) among haplotypes of P. glauca collected in Indonesia (Ind) and Australia (Aust). The phylogeny was rooted with C. falciformis and C. amblyrhynchos and nodal support is given as Bayesian posterior probabilities/ML boostrap support. Dash (-) indicates support of less than 50%. In the network, each indicated step (circle) represents a single nucleotide difference in the mtDNA control region sequence. The area of circles is scaled to represent the relative frequency of that haplotype and the smallest circle represent inferred haplotypes that were not sampled. The collection location of sampled haplotypes is numbered (in italics) according to Fig. 1.
Figure 5
Inferred phylogeny (A) and statistical parsimony network (B) among haplotypes of S. lewini. The phylogeny was rooted with S. mokarran and E. blochi and nodal support is given as Bayesian posterior probabilities/ML boostrap support. Dash (-) indicates support of less than 50%. In the network, each indicated step (circle) represents a single nucleotide difference in the mtDNA control region sequence. The area of circles is scaled to represent the relative frequency of that haplotype and the smallest circle represent inferred haplotypes that were not sampled. Haplotype SL09 from the North Atlantic (Atl) is equivalent to Duncan et al [15] haplotype number 16. The collection location of sampled haplotypes is numbered (in italics) according to Fig. 1.
Similar articles
- Genetic connectivity of the scalloped hammerhead shark Sphyrna lewini across Indonesia and the Western Indian Ocean.
Hadi S, Andayani N, Muttaqin E, Simeon BM, Ichsan M, Subhan B, Madduppa H. Hadi S, et al. PLoS One. 2020 Oct 1;15(10):e0230763. doi: 10.1371/journal.pone.0230763. eCollection 2020. PLoS One. 2020. PMID: 33002022 Free PMC article. - Conservation Genetics of the Scalloped Hammerhead Shark in the Pacific Coast of Colombia.
Quintanilla S, Gómez A, Mariño-Ramírez C, Sorzano C, Bessudo S, Soler G, Bernal JE, Caballero S. Quintanilla S, et al. J Hered. 2015;106 Suppl 1:448-58. doi: 10.1093/jhered/esv050. J Hered. 2015. PMID: 26245780 - Assessing multiple paternity in three commercially exploited shark species: Mustelus mustelus, Carcharhinus obscurus and Sphyrna lewini.
Rossouw C, Wintner SP, Bester-Van Der Merwe AE. Rossouw C, et al. J Fish Biol. 2016 Aug;89(2):1125-41. doi: 10.1111/jfb.12996. Epub 2016 May 29. J Fish Biol. 2016. PMID: 27237109 - Shark recreational fisheries: Status, challenges, and research needs.
Gallagher AJ, Hammerschlag N, Danylchuk AJ, Cooke SJ. Gallagher AJ, et al. Ambio. 2017 May;46(4):385-398. doi: 10.1007/s13280-016-0856-8. Epub 2016 Dec 19. Ambio. 2017. PMID: 27995551 Free PMC article. Review. - Shark depredation: future directions in research and management.
Mitchell JD, Drymon JM, Vardon J, Coulson PG, Simpfendorfer CA, Scyphers SB, Kajiura SM, Hoel K, Williams S, Ryan KL, Barnett A, Heupel MR, Chin A, Navarro M, Langlois T, Ajemian MJ, Gilman E, Prasky E, Jackson G. Mitchell JD, et al. Rev Fish Biol Fish. 2023;33(2):475-499. doi: 10.1007/s11160-022-09732-9. Epub 2022 Nov 15. Rev Fish Biol Fish. 2023. PMID: 36404946 Free PMC article. Review.
Cited by
- Population genetics of four heavily exploited shark species around the Arabian Peninsula.
Spaet JL, Jabado RW, Henderson AC, Moore AB, Berumen ML. Spaet JL, et al. Ecol Evol. 2015 Jun;5(12):2317-32. doi: 10.1002/ece3.1515. Epub 2015 May 20. Ecol Evol. 2015. PMID: 26120422 Free PMC article. - Decline of coastal apex shark populations over the past half century.
Roff G, Brown CJ, Priest MA, Mumby PJ. Roff G, et al. Commun Biol. 2018 Dec 13;1:223. doi: 10.1038/s42003-018-0233-1. eCollection 2018. Commun Biol. 2018. PMID: 30564744 Free PMC article. - Genetic differentiation and phylogeography of Mediterranean-North Eastern Atlantic blue shark (Prionace glauca, L. 1758) using mitochondrial DNA: panmixia or complex stock structure?
Leone A, Urso I, Damalas D, Martinsohn J, Zanzi A, Mariani S, Sperone E, Micarelli P, Garibaldi F, Megalofonou P, Bargelloni L, Franch R, Macias D, Prodöhl P, Fitzpatrick S, Stagioni M, Tinti F, Cariani A. Leone A, et al. PeerJ. 2017 Dec 6;5:e4112. doi: 10.7717/peerj.4112. eCollection 2017. PeerJ. 2017. PMID: 29230359 Free PMC article. - Brown banded bamboo shark (Chiloscyllium punctatum) shows high genetic diversity and differentiation in Malaysian waters.
Lim KC, Then AY, Wee AKS, Sade A, Rumpet R, Loh KH. Lim KC, et al. Sci Rep. 2021 Jul 21;11(1):14874. doi: 10.1038/s41598-021-94257-7. Sci Rep. 2021. PMID: 34290296 Free PMC article. - Crossing lines: a multidisciplinary framework for assessing connectivity of hammerhead sharks across jurisdictional boundaries.
Chin A, Simpfendorfer CA, White WT, Johnson GJ, McAuley RB, Heupel MR. Chin A, et al. Sci Rep. 2017 Apr 21;7:46061. doi: 10.1038/srep46061. Sci Rep. 2017. PMID: 28429742 Free PMC article.
References
- White WT, Last PR, JD S, Yearsley GK, Fahmi , Dharmadi . Economically important sharks and rays of Indonesia. Canberra, Australia, Australian Centre for International Agricultural Research; 2006.
- Blaber SJM, Dichmont CM, Buckworth RC, Badrudin , Sumiono B, Nurhakim S, Iskandar B, Fegan B, Ramm DC, Salini JP. Shared stocks of snappers (Lutjanidae) in Australia and Indonesia: Integrating biology, population dynamics and socio-economics to examine management scenarios. Reviews in Fish Biology and Fisheries. 2005;15:111–127. doi: 10.1007/s11160-005-3887-y. - DOI
- Last PR, Stevens JD. Sharks and Rays of Australia. CSIRO Australia. 1994.
- White WT, Last PR, Compagno LJV. Description of a new species of weasel shark, Hemigaleus australiensis n sp (Carcharhiniformes: Hemigaleidae) from Australian waters. Zootaxa. 2005. pp. 37–49.
- Stevens J, Simpfendorfer C, Francis M. Regional overview – southwest Pacific, Australasia and Oceania. In: Fowler S, Cavanagh R, Camhi M, Burgess G, Cailliet G, Fordham S, Simpfendorfer C, Musick J, editor. Sharks, Rays and Chimeras: The Status of the Chondrichthyan Fishes. Cambridge, UK: International Union for the Conservation of Nature and Natural Resources; 2005. pp. 161–171.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous