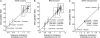Microsatellites are molecular clocks that support accurate inferences about history - PubMed (original) (raw)
Microsatellites are molecular clocks that support accurate inferences about history
James X Sun et al. Mol Biol Evol. 2009 May.
Abstract
Microsatellite length mutations are often modeled using the generalized stepwise mutation process, which is a type of random walk. If this model is sufficiently accurate, one can estimate the coalescence time between alleles of a locus after a mathematical transformation of the allele lengths. When large-scale microsatellite genotyping first became possible, there was substantial interest in using this approach to make inferences about time and demography, but that interest has waned because it has not been possible to empirically validate the clock by comparing it with data in which the mutation process is well understood. We analyzed data from 783 microsatellite loci in human populations and 292 loci in chimpanzee populations, and compared them with up to one gigabase of aligned sequence data, where the molecular clock based upon nucleotide substitutions is believed to be reliable. We empirically demonstrate a remarkable linearity (r(2) > 0.95) between the microsatellite average square distance statistic and sequence divergence. We demonstrate that microsatellites are accurate molecular clocks for coalescent times of at least 2 million years (My). We apply this insight to confirm that the African populations San, Biaka Pygmy, and Mbuti Pygmy have the deepest coalescent times among populations in the Human Genome Diversity Project. Furthermore, we show that microsatellites support unbiased estimates of population differentiation (F(ST)) that are less subject to ascertainment bias than single nucleotide polymorphism (SNP) F(ST). These results raise the prospect of using microsatellite data sets to determine parameters of population history. When genotyped along with SNPs, microsatellite data can also be used to correct for SNP ascertainment bias.
Figures
FIG. 1.—
Microsatellite ASD is linear with sequence divergence. Horizontal axes are sequence divergences measured in substitutions per kb, which we assume is an accurate gold standard. Vertical axes are microsatellite ASD values. Crosshairs are data with standard errors for each population pair. The linear regression line is shown. For WGS humans (A), the correlation coefficient is r = 0.989 (P = 4.9e−7, 95% CI 0.946–0.998). In the left box are Yoruba versus (top to bottom): European, East Asian, and Yoruba. In the right box are Biaka Pygmy versus (top to bottom): European, Yoruba, and East Asian. For RRS humans (B), r = 0.983 (P = 5.3e−11, 95% CI 0.949–0.995). In the left box are Yoruba versus (top to bottom): European, Australian Aborigine, East Asian, and Yoruba. In the right box is Biaka Pygmy versus: European, Yoruba, and East Asian; also are San versus: Yoruba, European, and East Asian. For chimpanzees (C), r = 0.986 (P = 2.7e−4, 95% CI 0.877–0.999).
FIG. 2.—
Inferred pairwise sequence divergences of HGDP populations. Microsatellite ASD for each pair of populations in HGDP is computed. Then using regression from figure 1_A_, we inferred the divergence of each population pair in substitutions per kb. The grayscale intensities display the range of divergences. As shown, San and Pygmy Africans are equidistant from all other populations, suggesting that they have the largest _t_MRCA to any other human population.
FIG. 3.—
Microsatellite and SNP _F_ST are almost equivalent, with the discrepancy likely due to SNP ascertainment. Horizontal axes are the SNP _F_ST. Vertical axes are the microsatellite _F_ST. In Panel A are _F_ST computed from real HGDP data. There are (53 choose 2) = 1,378 pairwise population comparisons (data points). Circles and plus signs are data for each population pair. The linearity is clear, and the regression lines (not shown) intersect the origin. However, there are two distinct slopes for _F_ST > 0.1. In circles are 1,035 (46 non-African populations, choose 2) non-Africans versus non-Africans, with regression line slope = 0.91 and correlation coefficient 0.983 (P < 1e−10, 95% CI 0.982–0.986). In plus signs are Africans versus all populations, with regression line slope = 0.73 and correlation coefficient 0.969 (P < 1e−10, 95% CI 0.962–0.975). In Panel B are simulated data (demographic model in
supplementary fig. S2_A_
,
Supplementary Material
online) with different SNP ascertainment schemes: No ascertainment in circles, ascertaining using two samples from population A (“African”) in dots, ascertaining using two samples from population B (“European”) in crosses, and ascertaining using one sample from each population in plus signs. In Panel C are simulated data (demographic model in
supplementary fig. S2_B_
,
Supplementary Material
online) of four populations resembling Africans, Europeans, East Asians, and Native Americans. We used the European-African ascertainment scheme (see text). In circles are non-Africans versus non-Africans. In plus signs are Africans versus non-Africans. For panels B and C, enough loci were simulated such that standard errors are of negligible magnitude.
FIG. 4.—
A unifying view of ASD and microsatellite _F_ST. The horizontal axis is interpopulation variance. The vertical axis is intrapopulation variance. Afr = Africans, NA = Native Americans, PI = Pacific Islanders, EA = East Asians, EMC = Europeans, Middle Easterners, and Central South Asians. It is shown (Materials and Methods) that microsatellite _F_ST and ASD are functions of these two variances. Lines of constant ASD are dashed lines with negative slope. Lines of constant _F_ST are dashed lines with positive slope. The data are (53 choose 2) = 1,378 pairwise HGDP population comparisons. Clearly, this picture segregates populations into distinguishable clusters. Africans versus all are above the thick black line. Non-Africans versus non-Africans are below the line. Distinguishable clusters are demarcated in ovals and squares.
Similar articles
- Factors influencing ascertainment bias of microsatellite allele sizes: impact on estimates of mutation rates.
Li B, Kimmel M. Li B, et al. Genetics. 2013 Oct;195(2):563-72. doi: 10.1534/genetics.113.154161. Epub 2013 Aug 14. Genetics. 2013. PMID: 23946335 Free PMC article. - Quantifying ascertainment bias and species-specific length differences in human and chimpanzee microsatellites using genome sequences.
Vowles EJ, Amos W. Vowles EJ, et al. Mol Biol Evol. 2006 Mar;23(3):598-607. doi: 10.1093/molbev/msj065. Epub 2005 Nov 21. Mol Biol Evol. 2006. PMID: 16301296 - Estimating genomic diversity and population differentiation - an empirical comparison of microsatellite and SNP variation in Arabidopsis halleri.
Fischer MC, Rellstab C, Leuzinger M, Roumet M, Gugerli F, Shimizu KK, Holderegger R, Widmer A. Fischer MC, et al. BMC Genomics. 2017 Jan 11;18(1):69. doi: 10.1186/s12864-016-3459-7. BMC Genomics. 2017. PMID: 28077077 Free PMC article. - Ascertainment bias cannot entirely account for human microsatellites being longer than their chimpanzee homologues.
Cooper G, Rubinsztein DC, Amos W. Cooper G, et al. Hum Mol Genet. 1998 Sep;7(9):1425-9. doi: 10.1093/hmg/7.9.1425. Hum Mol Genet. 1998. PMID: 9700197 - Divergent microsatellite evolution in the human and chimpanzee lineages.
Gáspári Z, Ortutay C, Tóth G. Gáspári Z, et al. FEBS Lett. 2007 May 29;581(13):2523-6. doi: 10.1016/j.febslet.2007.04.073. Epub 2007 May 4. FEBS Lett. 2007. PMID: 17498704
Cited by
- Unraveling the hierarchical genetic structure of tea green leafhopper, Matsumurasca onukii, in East Asia based on SSRs and SNPs.
Zhang L, Dietrich CH, Xu Y, Yang Z, Chen M, Pham TH, Le CCV, Qiao L, Matsumura M, Qin D. Zhang L, et al. Ecol Evol. 2022 Oct 1;12(10):e9377. doi: 10.1002/ece3.9377. eCollection 2022 Oct. Ecol Evol. 2022. PMID: 36203634 Free PMC article. - Estimations of Mutation Rates Depend on Population Allele Frequency Distribution: The Case of Autosomal Microsatellites.
Antão-Sousa S, Conde-Sousa E, Gusmão L, Amorim A, Pinto N. Antão-Sousa S, et al. Genes (Basel). 2022 Jul 14;13(7):1248. doi: 10.3390/genes13071248. Genes (Basel). 2022. PMID: 35886031 Free PMC article. - Origin and dispersion pathways of guava in the Galapagos Islands inferred through genetics and historical records.
Urquía D, Gutierrez B, Pozo G, Pozo MJ, Torres ML. Urquía D, et al. Ecol Evol. 2021 Oct 4;11(21):15111-15131. doi: 10.1002/ece3.8193. eCollection 2021 Nov. Ecol Evol. 2021. PMID: 34765164 Free PMC article. - Comparing the Performance of Microsatellites and RADseq in Population Genetic Studies: Analysis of Data for Pike (Esox lucius) and a Synthesis of Previous Studies.
Sunde J, Yıldırım Y, Tibblin P, Forsman A. Sunde J, et al. Front Genet. 2020 Mar 13;11:218. doi: 10.3389/fgene.2020.00218. eCollection 2020. Front Genet. 2020. PMID: 32231687 Free PMC article. - micRocounter: Microsatellite Characterization in Genome Assemblies.
Lo J, Jonika MM, Blackmon H. Lo J, et al. G3 (Bethesda). 2019 Oct 7;9(10):3101-3104. doi: 10.1534/g3.119.400335. G3 (Bethesda). 2019. PMID: 31375475 Free PMC article.
References
- Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, Lander ES. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. - PubMed
- Amos W, Rubinstzein DC. Microsatellites are subject to directional evolution. Nat Genet. 1996;12:13–14. - PubMed
- Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous