Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development - PubMed (original) (raw)
Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development
Hisanobu Oda et al. Mol Cell Biol. 2009 Apr.
Abstract
PR-Set7/Set8/KMT5A is the sole enzyme known to catalyze monomethylation of histone H4 lysine 20 (H4K20) and is present only in multicellular organisms that compact a large fraction of their DNA. We found that mouse embryos that are homozygous null mutants for the gene PR-Set7 display early embryonic lethality prior to the eight-cell stage. Death was due to the absence of PR-Set7 catalytic activity, since microinjection of the wild type, but not a catalytically inactive version, into two-cell embryos rescued the phenotype. A lack of PR-Set7 activity resulted not only in depletion of H4K20me1 but also in reduced levels of the H4K20me2/3 marks catalyzed by the Suv4-20h1/h2 enzymes, implying that H4K20me1 may be essential for the function of these enzymes to ensure the dimethylated and trimethylated states. Embryonic stem cells that were inducibly deleted for PR-Set7 passed through an initial G(2)/M phase, but the progeny were defective at the subsequent S and G(2)/M phases, exhibiting a delay in their cell cycle, accumulation at G(2)/M, massive DNA damage, and improper mitotic chromosome condensation. Cell cycle analysis after synchronization indicated that the defects were a consequence of decreased H4K20me1 due to the absence of PR-Set7. Most importantly, the lack of H4K20me1 also resulted in defects in chromosome condensation in interphase nuclei. These results demonstrate the critical role of H4K20 monomethylation in mammals in a developmental context.
Figures
FIG. 1.
Targeting strategy of PR-Set7 knockout mice. (A) Schematic representation of the targeting strategy to generate PR-Set7 knockout mice showing the C-terminal region of wt and genetically engineered alleles of PR-Set7. Exons are shown as green boxes, and loxP and FLP recombination target sites are shown as blue and red triangles, respectively. The exons encoding the SET domain, the relevant restriction sites (XbaI and EcoRI), the sizes of the restriction fragments used for genotyping, and the position of the probes are indicated. The floxed allele functions as the wt and contains two loxP sites flanking exon 7 that include the beginning of the SET domain. Cre-mediated and FLPe-mediated recombination of targeted alleles results in a null allele and a floxed allele, respectively. HSV-tk, herpes simplex virus thymidine kinase. (B) Southern blotting to confirm homologous recombination in two ES clones (68 and 189) used for chimera production. Genomic DNA was digested with EcoRI and hybridized with 3′ probe as indicated in panel A. (C) PCR genotyping of PR-Set7 wt (+/+) and heterozygous (+/−) mice.
FIG. 2.
PR-Set7 knockout embryos arrest at late G2 or M phase prior to the eight-cell stage and exhibit decreased H4K20me1, increased H3S10P, and apoptosis. (A) Microscopic analysis of embryos at 2.5 dpc that were collected and transferred to in vitro culture for a day. (Top) Embryos having eight or more cells developed into blastocysts after 1 day of culture. (Bottom) Those having less than eight cells did not develop into blastocysts and degenerated. (B) Genotyping results of in vitro-cultured embryos shown in panel A. Embryos that developed into blastocysts were wt (+/+) or heterozygous (+/−). All the embryos that had less than eight cells at 2.5 dpc were homozygous (−/−). (C) Immunofluorescence analysis using anti-H4K20me1 and anti-H3S10P antibodies. (Left) A 2.5-dpc embryo wt for PR-Set7 exhibits variable levels of H4K20me1 in each blastomere, and the H3S10P signal is negative. (Right) PR-Set7 KO embryos arrest with nearly undetectable levels of H4K20me1 in the blastomeres but with clear staining in the polar body (PB; arrow). H3S10P shows speckled or diffuse staining in all blastomeres. (D) TUNEL assay performed on 2.5-dpc embryos after in vitro culture for a day. Presumptive PR-Set7 KO embryos showed positive signals (white arrows) as opposed to other embryos that showed few, weak signals and developed into blastocysts. (E) Comparison of homozygous null (_PR-Set7_−/−) versus wt (PR-Set7+/+) 2.5-dpc embryos in an in situ caspase assay after in vitro culture for a day. The _PR-Set7_−/− embryo shows strong caspase-3 and caspase-7 activity.
FIG. 3.
Time-lapse analysis of embryos derived from PR-Set7+/− crosses. Embryos were obtained 44 h post-human chorionic gonadotropin injection from 6-week-old females. Two-cell-stage embryos were cultured in KSOM covered with paraffin oil in a 37°C chamber under a 5% CO2 atmosphere. Embryos were imaged under differential interference contrast optics with the ×20 objective of an inverted Leica microscope. Images were captured every 20 min in different z planes (covering 60 μm) and in four different fields using an automated stage controller.
FIG. 4.
The methyltransferase activity of PR-Set7 is required to rescue the knockout embryo. (A) Schematic representation of HA-tagged PR-Set7, either wt or mutant, in the catalytic SET domain. The experimental strategy for microinjection of mRNAs encoding GFP- and HA-tagged PR-Set7, either wt or mutant (mut), into one cell of a two-cell embryo is shown. The embryos were derived from intercrossing mice heterozygous for the PR-Set7 null allele. (B) Bright-field microscopy and fluorescence analyses for GFP expression of embryos microinjected with the mRNAs are indicated to the left. Injection of wt but not mutant PR-Set7 mRNA into one cell of a two-cell-stage embryo rescues the injected cells in PR-Set7 null embryos. Note that in these rescued embryos, cell proliferation and in some instances avitation are observed by day 3 (bottom, black arrows). (C) As described for panel B, a blastomere corresponding to the injected side of a two-cell-stage embryo that was presumably _PR-Set7_−/− developed into a blastocyst-like structure and exhibited formation of a blastocoelic cavity. The white broken line shows an uninjected blastomere(s) (e.g., GFP negative), which is presumably degenerated by day 3. (D) Analysis of an equivalent to an approximately eight-cell-stage embryo after injection of one cell of a two-cell embryo with wt PR-Set7 and GFP mRNAs. The panels show DAPI staining, indirect immunofluorescence using anti-H4K20me1 antibodies, and direct fluorescence from GFP expression. The last indicates the progeny of the injected blastomere (dashed white line). H4K20me1 appears distributed throughout the nuclei of the injected cells (dashed line; the arrow points to the polar body [PB]) but not in the other half of the embryo. The reduced H4K20me1 levels and DNA fragmentation observed in this other half indicate that the embryo is _PR-Set7_−/−. In contrast to that of noninjected cells, the nuclear morphology of the injected cells seemed normal. (E) Single-embryo genotyping of the embryo shown in panel D confirmed it to be _PR-Set7_−/−. PR-Set7+/+ and PR-Set7+/− genotypes are shown as controls on the right.
FIG. 5.
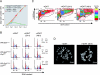
Marked changes in H4K20me1 and PR-Set7 expression at G2/M. (A) FACS analysis of asynchronous or NCZ-arrested F9 cells as indicated. The G1, S, and G2/M populations are indicated. Each panel plots the fluorescent signal intensity (log scale) of the H4K20me1 antibody in arbitrary units against DNA content in arbitrary units. (B) (Top) Western analysis of histone H3 and H4 modifications and PR-Set7 protein in synchronized HeLa cells using the antibodies indicated to the left. GAPDH provides a loading control. Cells released from G1 arrest after a double-thymidine block were analyzed every 2 h as indicated at the top. (Bottom) Cell cycle phases were confirmed by FACS analyses, and the data are represented by a histogram.
FIG. 6.
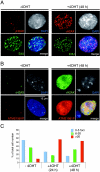
The levels of all three methylation states of H4K20 decrease in PR-Set7 conditional knockout ES cells. (A) Genotype analysis of PR-Set7 conditional knockout ES cells (see Materials and Methods) as a function of time of CreERT activation with 4-OHT as indicated at the top (see text). Both the XEN cell line that has PR-Set7 flox/+ but lacks Cre ERT and PGK12.1 ES cells were used as negative controls for 4-OHT treatment. The expected migration of DNA fragments containing floxed, wt, or null PR-Set7 alleles is shown to the left. PR-Set7+/− and PR-Set7_−/− ES cells were derived from IKE5-1 (PR-Set7 flox/+; Cre_ ERT) and IKE5-2 (PR-Set7 flox/−; Cre ERT) ES cells, respectively, after conditional knockout of PR-Set7 during 2 days of 4-OHT treatment. (B) Western blot analyses using the antibodies are shown to the left, and nuclear protein isolated from the cells indicated as a function of time of 4-OHT treatment is also indicated at the top. The levels of H4K20me1, H4K20me2, and H4K20me3 declined appreciably in the case of _PR-Set7_−/− ES cells, derived by treating PR-Set7 flox/−; Cre ERT ES cells with 4-OHT for 2 days. (C, D, and E) Immunofluorescence analysis using anti-H4K20me1 antibody (C) or anti-H4K20me3 antibody (D and E) and PR-Set7 flox/−; Cre ERT cells either untreated or treated with 4-OHT for 1 or 2 days as indicated to the left.
FIG. 7.
Conditional PR-Set7 KO ES cells accumulate at G2/M phase. (A) Growth curve for PR-Set7 flox/−_; Cre_ ERT and PR-Set7+/+ as a function of 4-OHT treatment as indicated. PR-Set7 flox/−_; Cre_ ERT cells showed a slow growth in the presence of 4-OHT, whereas the cells continued to proliferate in its absence. (B) FACS analyses of the cells are indicated to the left as a function of the number of days of 4-OHT treatment as indicated at the top. Conditional knockout of PR-Set7 generated by 2 days of 4-OHT treatment of PR-Set7 flox/−_; Cre_ ERT cells led to significant accumulation at the G2/M phase (bottom right). (C) The fluorescent signal intensity (log scale) of the antibody against H4K20me1 in arbitrary units is plotted against the DNA content in arbitrary units for PR-Set7 flox/−; Cre ERT cells treated with 4-OHT for the number of days indicated above each plot. Six color divisions represent the plot density from highest to lowest as follows: yellow, magenta, blue, blue-green, green, and red. The highest density area in each panel is indicated by an arrow. (D) Morphology of chromosome in PR-Set7 flox/−_; Cre_ ERT cells without 4-OHT treatment (left) and with 4-OHT treatment (right) for 2 days. (Right) The arrowhead shows separated sister centromeres. −4OHT, without 4-OHT treatment; +40HT, with 4-OHT treatment.
FIG. 8.
Cell cycle analysis after PR-Set7 depletion. (A) Experimental design. PR-Set7 flox/−; Cre ERT cells were synchronized with NCZ for 12 h followed by treatment with thymidine in the presence or absence of 4-OHT for 24 h and released into fresh ES medium. Samples were collected for FACS analysis and protein extraction at the time points shown at the top (black triangles). (B) Changes of the level of PR-Set7 protein and H4K20me1. Shown are Western blot analyses using the antibodies shown to the left and nuclear protein isolated from the cells at the time points indicated at the top. PR-Set7 protein was undetectable at 0 h after 24 h of treatment with 4-OHT (bottom). Both β-tubulin and histone H4 are shown as loading controls. (C) FACS analyses of the cells at each time point as indicated at the top. Note that cells treated with 4-OHT (bottom) show slower cell cycle progression between 8 h and 12 h than the cells untreated with 4-OHT (top) and accumulate at early S phase at 12 h (bottom right). AS, asynchronous.
FIG. 9.
DNA synthesis decreases during S phase, and γ-H2AX foci increase after conditional knockout of PR-Set7 in ES cells. (A and B) Immunofluorescence analyses using untreated PR-Set7 flox/−; Cre ERT cells (−4OHT) or those exposed to 4-OHT for 48 h [+4OHT (48 h)] as indicated at the top. Colocalization of antibody specific to γ-H2AX with either EdU or antibody specific to phospho-ATM is shown (merge). Phosphorylation of γ-H2AX at serine 139 and of the ATM kinase at serine 1981 occurs in response to DNA damage. (C) The percentage of PR-Set7 flox/−; Cre ERT cells exhibiting the indicated number of foci of colocalized γ-H2AX and phospho-ATM as a function of 4-OHT treatment for the number of hours shown at the bottom. The number of colocalized foci increased dramatically after knockout of PR-Set7.
FIG. 10.
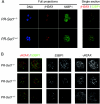
Analysis of 53BP1 and γ-H2AX in embryos from PR-Set7+/− intercrosses. (A) Embryos from PR-Set7+/− intercrosses were recovered between the four- and the eight-cell stage and immediately processed for immunostaining with antibodies against 53BP1 and γ-H2AX. Shown are full projections of stack sections taken every 1 μm or merged single-section images as indicated. Note that 53BP1 displays both cytoplasmic and nuclear localization. After imaging, embryos were genotyped individually as indicated. (B) Single sections at a higher magnification of two representative nuclei from either PR-Set7 null, wt, or heterozygous embryos are shown. No major differences in levels of either 53BP1 or γ-H2AX were detected between PR-Set7 heterozygous embryos, wt embryos, and null embryos.
FIG. 11.
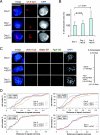
Increased chromosomal territory area and intergenic distances indicative of global chromosomal decondensation following conditional knockout of PR-Set7 in ES cells. (A) DNA FISH using an X chromosome paint probe on PR-Set7 flox/−; Cre ERT cells after 0, 1, and 2 days of 4-OHT treatment. Bar, 10 μm. (B) Quantification of relative chromosome area for each time point. The area of the X chromosome in z projections of 3D stacks was measured and normalized to the nuclear area for 50 nuclei at each point of 4-OHT treatment. Significance values are shown at the top. (C) (Left) Triple-probe DNA FISH using BAC probes for the X-linked genes Ureb1 (Cy5), G6pdx (Spectrum Red [SR]), and Fgd1 (Spectrum Green [SG]) following 0, 1, and 2 days of 4-OHT treatment of PR-Set7 flox/−; Cre ERT ES cells. DAPI results are shown in blue. Maximum intensity projections of 3D image stacks (five to seven planes) are shown. (Right) Map of the X chromosome showing the positions of the genes tested. (D) PR-Set7 flox/−; Cre ERT cells show an increase in intergenic distances after 4-OHT treatment. Intergenic distances for each DNA FISH probe are shown following day 0, 1, and 2 for 4-OHT-treated PR-Set7 flox/−; Cre ERT cells and for the G6pdx-Ureb1 probes for day 0 and 2 for 4-OHT-treated PR-Set7 flox/+; Cre ERT cells. Distances were measured between the two centers of mass of the DNA FISH signals from 3D stacks of images and then normalized to the nuclear volume using an ImageJ plug-in (n = 30 per day of treatment). +4OHT, 4-OHT treated.
Similar articles
- Methylation of histone H4 lysine 20 by PR-Set7 ensures the integrity of late replicating sequence domains in Drosophila.
Li Y, Armstrong RL, Duronio RJ, MacAlpine DM. Li Y, et al. Nucleic Acids Res. 2016 Sep 6;44(15):7204-18. doi: 10.1093/nar/gkw333. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131378 Free PMC article. - PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription.
Beck DB, Oda H, Shen SS, Reinberg D. Beck DB, et al. Genes Dev. 2012 Feb 15;26(4):325-37. doi: 10.1101/gad.177444.111. Genes Dev. 2012. PMID: 22345514 Free PMC article. Review. - A new regulator of the cell cycle: the PR-Set7 histone methyltransferase.
Wu S, Rice JC. Wu S, et al. Cell Cycle. 2011 Jan 1;10(1):68-72. doi: 10.4161/cc.10.1.14363. Epub 2011 Jan 1. Cell Cycle. 2011. PMID: 21200139 Free PMC article. Review. - Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability.
Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC. Houston SI, et al. J Biol Chem. 2008 Jul 11;283(28):19478-88. doi: 10.1074/jbc.M710579200. Epub 2008 May 14. J Biol Chem. 2008. PMID: 18480059 Free PMC article. - The histone H4 lysine 20 monomethyl mark, set by PR-Set7 and stabilized by L(3)mbt, is necessary for proper interphase chromatin organization.
Sakaguchi A, Joyce E, Aoki T, Schedl P, Steward R. Sakaguchi A, et al. PLoS One. 2012;7(9):e45321. doi: 10.1371/journal.pone.0045321. Epub 2012 Sep 14. PLoS One. 2012. PMID: 23024815 Free PMC article.
Cited by
- Methylation of histone H4 lysine 20 by PR-Set7 ensures the integrity of late replicating sequence domains in Drosophila.
Li Y, Armstrong RL, Duronio RJ, MacAlpine DM. Li Y, et al. Nucleic Acids Res. 2016 Sep 6;44(15):7204-18. doi: 10.1093/nar/gkw333. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131378 Free PMC article. - Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity.
Jørgensen S, Schotta G, Sørensen CS. Jørgensen S, et al. Nucleic Acids Res. 2013 Mar 1;41(5):2797-806. doi: 10.1093/nar/gkt012. Epub 2013 Jan 23. Nucleic Acids Res. 2013. PMID: 23345616 Free PMC article. Review. - Epigenetic and Transcriptional Control of Erythropoiesis.
Wells M, Steiner L. Wells M, et al. Front Genet. 2022 Mar 7;13:805265. doi: 10.3389/fgene.2022.805265. eCollection 2022. Front Genet. 2022. PMID: 35330735 Free PMC article. Review. - Multiple distinct domains of human XIST are required to coordinate gene silencing and subsequent heterochromatin formation.
Dixon-McDougall T, Brown CJ. Dixon-McDougall T, et al. Epigenetics Chromatin. 2022 Feb 4;15(1):6. doi: 10.1186/s13072-022-00438-7. Epigenetics Chromatin. 2022. PMID: 35120578 Free PMC article. - CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase.
Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L. Centore RC, et al. Mol Cell. 2010 Oct 8;40(1):22-33. doi: 10.1016/j.molcel.2010.09.015. Mol Cell. 2010. PMID: 20932472 Free PMC article.
References
- Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. - PubMed
- Dorigo, B., T. Schalch, K. Bystricky, and T. J. Richmond. 2003. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 32785-96. - PubMed
- Fraga, M. F., E. Ballestar, A. Villar-Garea, M. Boix-Chornet, J. Espada, G. Schotta, T. Bonaldi, C. Haydon, S. Ropero, K. Petrie, N. G. Iyer, A. Perez-Rosado, E. Calvo, J. A. Lopez, A. Cano, M. J. Calasanz, D. Colomer, M. A. Piris, N. Ahn, A. Imhof, C. Caldas, T. Jenuwein, and M. Esteller. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37391-400. - PubMed
- Hayashi, S., P. Lewis, L. Pevny, and A. P. McMahon. 2002. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119(Suppl. 1)S97-S101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous