HPC2 and ubinuclein define a novel family of histone chaperones conserved throughout eukaryotes - PubMed (original) (raw)
HPC2 and ubinuclein define a novel family of histone chaperones conserved throughout eukaryotes
S Balaji et al. Mol Biosyst. 2009 Mar.
Abstract
While histone chaperones have been intensely studied, the roles of components of the Hir-Asf1 histone chaperone complex such as Hir3p and Hpc2p are poorly understood. Using sensitive protein sequence profile analyses we investigated the evolution of these proteins and showed that Hir3p and Hpc2p have a much wider phyletic pattern than was previously known. We established the animal histone-deacetylase-complex-interacting proteins, CAIN/CABIN, to be orthologs of Hir3p. They contain a conserved core of around 30 TPR-like bi-helical repeats that are likely to form a super-helical scaffold. We identified a conserved domain, the HUN domain, in all Hpc2p homologs, including animal ubinuclein/yemanuclein and the recently discovered vertebrate cell-cycle regulator FLJ25778. The HUN domain has a characteristic pattern of conserved acidic residues based on which we predict that it is a previously unrecognized histone-tail-binding chaperone. By analyzing various high-throughput data sets, such as RNAi knock-downs, genetic and protein interaction maps and cell-cycle-specific gene expression data, we present evidence that Hpc2p homologs might be deployed in specific processes of chromatin dynamics relating to cell-cycle progression in vertebrates and schizogony in Plasmodium. Beyond the conserved HUN domain these proteins show extensive divergence patterns in different eukaryotic lineages. Hence, we propose that Hpc2p homologs are probably involved in recruitment of the ancient conserved histone-loading Hir-Asf1 complex to different lineage-specific chromatin reorganization processes.
Figures
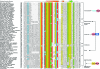
Fig. 1. Multiple sequence alignment and domain architectures of the HUN domain. Proteins are denoted by their gene name, species abbreviations and gi. Amino acids are colored based on their functional properties and conservation (>80%) in the alignment. The predicted secondary structure is shown at the top with α-helices represented as orange cylinders and β-strands as green block arrows. Domain architectures of HUN containing protein are shown to the right. Consensus abbreviations and coloring scheme are as follows: polar (p: QNSTCHKRDE) residues shaded blue, hydrophobic (h: ACFMWYIVL), aliphatic (l: IVL) and aromatic (a: FYWH) residues colored yellow, acidic (‘-’: DE) residues shaded red, small (s: GASVTDNPC) and tiny (u: GAS) residues colored green. Species abbreviations are as follows: Acap: Ajellomyces capsulatus; Afum: Aspergillus fumigatus; Agam: Anopheles gambiae; Amel: Apis mellifera; Anid: Aspergillus nidulans; Anig: Aspergillus niger; Aory: Aspergillus oryzae; Atha: Arabidopsis thaliana; Bbov: Babesia bovis; Bfuc: Botryotinia fuckeliana; Bmal: Brugia malayi; Calb: Candida albicans; Ccin: Coprinopsis cinerea; Cgla: Candida glabrata; Cglo: Chaetomium globosum; Cneo: Cryptococcus neoformans; Cpar: Cryptosporidium parvum; Crei: Chlamydomonas reinhardtii; Ddis: Dictyostelium discoideum; Dmel: Drosophila melanogaster; Dpse: Drosophila pseudoobscura; Drer: Danio rerio; Ecun: Encephalitozoon cuniculi; Ehis: Entamoeba histolytica; Ggal: Gallus gallus; Gzea: Gibberella zeae; Hsap: Homo sapiens; Klac: Kluyveromyces lactis; Mdom: Monodelphis domestica; Mglo: Malassezia globosa; Mgri: Magnaporthe grisea; Mmus: Mus musculus; Ncra: Neurospora crassa; Nvec: Nematostella vectensis; Nvit: Nasonia vitripennis; Osat: Oryza sativa; Otau: Ostreococcus tauri; Pfal: Plasmodium falciparum; Pnod: Phaeosphaeria nodorum; Ppat: Physcomitrella patens; Ptet: Paramecium tetraurelia; Pviv: Plasmodium vivax; Scer: Saccharomyces cerevisiae; Spom: Schizosaccharomyces pombe; Spur: Strongylocentrotus purpuratus; Tadh: Trichoplax adhaerens; Tann: Theileria annulata; Tnig: Tetraodon nigroviridis; Tpar: Theileria parva; Tthe: Tetrahymena thermophila; Tvag: Trichomonas vaginalis; Umay: Ustilago maydis; Vvin: Vitis vinifera; X(Si): Xenopus (Silurana); Ylip: Yarrowia lipolytica.
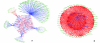
Fig. 2. (A) A network representation of genetic and protein interactions of Hir complex components (Hir1, Hir2, Hir3 and Hpc2) and Asf1. Nodes and edges in the network, respectively, represent genes/proteins and interactions between them. The shared interactions between two or more of the components of the Hir complex are shown within the central pentagon module, while the unique interactions of the components (that are not shared with other components of the Hir complex) are shown outside of the pentagon module. Asf1 having functions independent of the Hir complex (e.g. the Caf-1 complex) has a large set of unique interactions. Histones H2A, H3, H4, H2AZ are shown as purple nodes within the central pentagon module and components of the mediator/CDK8 complex are shown as green nodes (Med1, Med2, Ssn2, Cse2, Ssn8 and Med11) within the module. (B) Largest dense sub-network of proteins containing HUN domain protein FLJ25778 constructed based on mutual information in their phyletic patterns. The network has 64 human proteins implicated in cell-cycle function via RNAi knockdowns, which also contains FLJ25778, are shown as nodes. These nodes are connected by an edge if a given protein pair has a high MIC (> 0.6) in their phyletic patterns (see section 4.1 in ESI for details) and there are 908 edges in the network. The edges connecting protein FLJ25778 to other proteins in the network are shown in blue; among them transcription factors are shown as yellow nodes and the others are shown as blue nodes.
Similar articles
- Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells.
Banumathy G, Somaiah N, Zhang R, Tang Y, Hoffmann J, Andrake M, Ceulemans H, Schultz D, Marmorstein R, Adams PD. Banumathy G, et al. Mol Cell Biol. 2009 Feb;29(3):758-70. doi: 10.1128/MCB.01047-08. Epub 2008 Nov 24. Mol Cell Biol. 2009. PMID: 19029251 Free PMC article. - Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex.
Rai TS, Puri A, McBryan T, Hoffman J, Tang Y, Pchelintsev NA, van Tuyn J, Marmorstein R, Schultz DC, Adams PD. Rai TS, et al. Mol Cell Biol. 2011 Oct;31(19):4107-18. doi: 10.1128/MCB.05546-11. Epub 2011 Aug 1. Mol Cell Biol. 2011. PMID: 21807893 Free PMC article. - Functional Analysis of Hif1 Histone Chaperone in Saccharomyces cerevisiae.
Dannah NS, Nabeel-Shah S, Kurat CF, Sabatinos SA, Fillingham J. Dannah NS, et al. G3 (Bethesda). 2018 May 31;8(6):1993-2006. doi: 10.1534/g3.118.200229. G3 (Bethesda). 2018. PMID: 29661843 Free PMC article. - Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes.
Iyer LM, Anantharaman V, Wolf MY, Aravind L. Iyer LM, et al. Int J Parasitol. 2008 Jan;38(1):1-31. doi: 10.1016/j.ijpara.2007.07.018. Epub 2007 Sep 15. Int J Parasitol. 2008. PMID: 17949725 Review. - Histone chaperones, a supporting role in the limelight.
Loyola A, Almouzni G. Loyola A, et al. Biochim Biophys Acta. 2004 Mar 15;1677(1-3):3-11. doi: 10.1016/j.bbaexp.2003.09.012. Biochim Biophys Acta. 2004. PMID: 15020040 Review.
Cited by
- The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3.
Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. Drané P, et al. Genes Dev. 2010 Jun 15;24(12):1253-65. doi: 10.1101/gad.566910. Epub 2010 May 26. Genes Dev. 2010. PMID: 20504901 Free PMC article. - DAXX-dependent supply of soluble (H3.3-H4) dimers to PML bodies pending deposition into chromatin.
Delbarre E, Ivanauskiene K, Küntziger T, Collas P. Delbarre E, et al. Genome Res. 2013 Mar;23(3):440-51. doi: 10.1101/gr.142703.112. Epub 2012 Dec 5. Genome Res. 2013. PMID: 23222847 Free PMC article. - Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex.
Ricketts MD, Frederick B, Hoff H, Tang Y, Schultz DC, Singh Rai T, Grazia Vizioli M, Adams PD, Marmorstein R. Ricketts MD, et al. Nat Commun. 2015 Jul 10;6:7711. doi: 10.1038/ncomms8711. Nat Commun. 2015. PMID: 26159857 Free PMC article. - A single mutation results in diploid gamete formation and parthenogenesis in a Drosophila yemanuclein-alpha meiosis I defective mutant.
Meyer RE, Delaage M, Rosset R, Capri M, Aït-Ahmed O. Meyer RE, et al. BMC Genet. 2010 Nov 16;11:104. doi: 10.1186/1471-2156-11-104. BMC Genet. 2010. PMID: 21080953 Free PMC article. - The histone variant H3.3 claims its place in the crowded scene of epigenetics.
Bano D, Piazzesi A, Salomoni P, Nicotera P. Bano D, et al. Aging (Albany NY). 2017 Mar 10;9(3):602-614. doi: 10.18632/aging.101194. Aging (Albany NY). 2017. PMID: 28284043 Free PMC article. Review.
References
- Loyola A., Almouzni G. Biochim. Biophys. Acta. 2004;1677:3–11. - PubMed
- Henikoff S. Nat. Rev. Genet. 2008;9:15–26. - PubMed
- Akey C. W., Luger K. Curr. Opin. Struct. Biol. 2003;13:6–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources