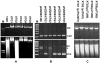DNA phosphorothioation in Streptomyces lividans: mutational analysis of the dnd locus - PubMed (original) (raw)
DNA phosphorothioation in Streptomyces lividans: mutational analysis of the dnd locus
Tiegang Xu et al. BMC Microbiol. 2009.
Abstract
Background: A novel DNA phosphorothioate modification (DNA sulfur modification), in which one of the non-bridging oxygen atoms in the phosphodiester bond linking DNA nucleotides is exchanged by sulphur, was found to be genetically determined by dnd or dnd-counterpart loci in a wide spectrum of bacteria from diverse habitats. A detailed mutational analysis of the individual genes within the dnd locus in Streptomyces lividans responsible for DNA phosphorothioation was performed and is described here. It should be of great help for the mechanistic study of this intriguing system.
Results: A 6,665-bp DNA region carrying just five ORFs (dndA-E) was defined as the sole determinant for modification of the DNA backbone in S. lividans to form phosphorothioate. This provides a diagnostically reliable and easily assayable Dnd (DNA degradation) phenotype. While dndA is clearly transcribed independently, dndB-E constitute an operon, as revealed by RT-PCR analysis. An efficient mutation-integration-complementation system was developed to allow for detailed functional analysis of these dnd genes. The Dnd- phenotype caused by specific in-frame deletion of the dndA, C, D, and E genes or the enhanced Dnd phenotype resulting from in-frame deletion of dndB could be restored by expression vectors carrying the corresponding dnd genes. Interestingly, overdosage of DndC or DndD, but not other Dnd proteins, in vivo was found to be detrimental to cell viability.
Conclusion: DNA phosphorothioation is a multi-enzymatic and highly coordinated process controlled by five dnd genes. Overexpression of some proteins in vivo prevented growth of host strain, suggesting that expression of the gene cluster is strictly regulated in the native host.
Figures
Figure 1
Localization of the boundaries for dnd gene cluster. pSET152-derivatives with the ability to confer Dnd (+ or -) phenotypes are indicated in line with their insert fragments. Five arrows from left to right represent five the ORFs of the dnd gene cluster (dndA-E). Directions of the arrows indicate the transcriptional directions of the genes.
Figure 2
RT-PCR analysis of the dnd genes transcripts. dnd gene transcripts were reverse transcribed and amplified. (A) Relative positions and directions of corresponding primers are marked with black arrows. (B) Amplification products with sense primer (SP), anti sense primer (AP) and their corresponding lengths. Intra-dnd gene amplification products are indicated as dnd gene names, while products of regions between dnd genes are named linking two corresponding genes such as AB. Amplification of 16S rRNA is used as an internal control marker (IM). (C) Electrophoresis of RT-PCR products. The amplification products are labeled as in Figure 2B. Reverse transcriptase inactivation (BC*) and without DNase treatment (AB') were carried out as negative and positive controls. DNA markers are labeled as "M".
Figure 3
dnd mutants. Black arrows represent dnd genes and their transcriptional directions. White blocks/arrows represent in-frame deletions in the corresponding genes.
Figure 4
Dnd phenotype of 1326 and related dnd mutants. (A) Dnd phenotype of chromosomal DNA for 1326 and related dnd mutants. (B) Dnd phenotype of plasmids pHZ209 isolated from 1326 and related dnd mutants. (C) Dnd phenotype of chromosomal DNA from complemented dnd mutants. DNA was first treated with TAE (top panel) or peracetic acid TAE (bottom panel) before fractionation by electrophoresis in TAE with added thiourea. M: DNA markers; CCC: covalently closed circular plasmid; OC: open circular plasmid. L: linear plasmid.
Figure 5
Western blotting for detecting expression of Dnd proteins in S. lividans 1326 and derivative strains. Rabbit polyclonal antibody to DndD reacted with the protein extracted from wild-type S. lividans 1326 or strain XTG4/pJTU64 (a pHZ1272-derived dndD expression vector).
Similar articles
- Twenty years hunting for sulfur in DNA.
Chen S, Wang L, Deng Z. Chen S, et al. Protein Cell. 2010 Jan;1(1):14-21. doi: 10.1007/s13238-010-0009-y. Epub 2010 Feb 7. Protein Cell. 2010. PMID: 21203994 Free PMC article. Review. - A novel DNA modification by sulphur.
Zhou X, He X, Liang J, Li A, Xu T, Kieser T, Helmann JD, Deng Z. Zhou X, et al. Mol Microbiol. 2005 Sep;57(5):1428-38. doi: 10.1111/j.1365-2958.2005.04764.x. Mol Microbiol. 2005. PMID: 16102010 - Phosphorothioation of DNA in bacteria by dnd genes.
Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, Zhou X, You D, Deng Z, Dedon PC. Wang L, et al. Nat Chem Biol. 2007 Nov;3(11):709-10. doi: 10.1038/nchembio.2007.39. Epub 2007 Oct 14. Nat Chem Biol. 2007. PMID: 17934475 - A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans.
You D, Wang L, Yao F, Zhou X, Deng Z. You D, et al. Biochemistry. 2007 May 22;46(20):6126-33. doi: 10.1021/bi602615k. Epub 2007 May 1. Biochemistry. 2007. PMID: 17469805 - dndDB: a database focused on phosphorothioation of the DNA backbone.
Ou HY, He X, Shao Y, Tai C, Rajakumar K, Deng Z. Ou HY, et al. PLoS One. 2009;4(4):e5132. doi: 10.1371/journal.pone.0005132. Epub 2009 Apr 9. PLoS One. 2009. PMID: 19357771 Free PMC article.
Cited by
- Crystal structure of the cysteine desulfurase DndA from Streptomyces lividans which is involved in DNA phosphorothioation.
Chen F, Zhang Z, Lin K, Qian T, Zhang Y, You D, He X, Wang Z, Liang J, Deng Z, Wu G. Chen F, et al. PLoS One. 2012;7(5):e36635. doi: 10.1371/journal.pone.0036635. Epub 2012 May 3. PLoS One. 2012. PMID: 22570733 Free PMC article. - Novel Iodine-induced Cleavage Real-time PCR Assay for Accurate Quantification of Phosphorothioate Modified Sites in Bacterial DNA.
Chen Y, Zheng T, Li J, Cui J, Deng Z, You D, Yang L. Chen Y, et al. Sci Rep. 2019 May 16;9(1):7485. doi: 10.1038/s41598-019-44011-x. Sci Rep. 2019. PMID: 31097783 Free PMC article. - Shewanella decolorationis LDS1 Chromate Resistance.
Lemaire ON, Honoré FA, Tempel S, Fortier EM, Leimkühler S, Méjean V, Iobbi-Nivol C. Lemaire ON, et al. Appl Environ Microbiol. 2019 Aug 29;85(18):e00777-19. doi: 10.1128/AEM.00777-19. Print 2019 Sep 15. Appl Environ Microbiol. 2019. PMID: 31300400 Free PMC article. - Metabolic and evolutionary insights into the closely-related species Streptomyces coelicolor and Streptomyces lividans deduced from high-resolution comparative genomic hybridization.
Lewis RA, Laing E, Allenby N, Bucca G, Brenner V, Harrison M, Kierzek AM, Smith CP. Lewis RA, et al. BMC Genomics. 2010 Dec 1;11:682. doi: 10.1186/1471-2164-11-682. BMC Genomics. 2010. PMID: 21122120 Free PMC article. - Twenty years hunting for sulfur in DNA.
Chen S, Wang L, Deng Z. Chen S, et al. Protein Cell. 2010 Jan;1(1):14-21. doi: 10.1007/s13238-010-0009-y. Epub 2010 Feb 7. Protein Cell. 2010. PMID: 21203994 Free PMC article. Review.
References
- Swinton D, Hattman S, Crain PF, Cheng CS, Smith DL, McCloskey JA. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2'-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc Natl Acad Sci USA. 1983;80(24):7400–7404. doi: 10.1073/pnas.80.24.7400. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases