Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants - PubMed (original) (raw)
Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants
Martin F Arlt et al. Am J Hum Genet. 2009 Mar.
Abstract
Copy number variants (CNVs) are an important component of genomic variation in humans and other mammals. Similar de novo deletions and duplications, or copy number changes (CNCs), are now known to be a major cause of genetic and developmental disorders and to arise somatically in many cancers. A major mechanism leading to both CNVs and disease-associated CNCs is meiotic unequal crossing over, or nonallelic homologous recombination (NAHR), mediated by flanking repeated sequences or segmental duplications. Others appear to involve nonhomologous end joining (NHEJ) or aberrant replication suggesting a mitotic cell origin. Here we show that aphidicolin-induced replication stress in normal human cells leads to a high frequency of CNCs of tens to thousands of kilobases across the human genome that closely resemble CNVs and disease-associated CNCs. Most deletion and duplication breakpoint junctions were characterized by short (<6 bp) microhomologies, consistent with the hypothesis that these rearrangements were formed by NHEJ or a replication-coupled process, such as template switching. This is a previously unrecognized consequence of replication stress and suggests that replication fork stalling and subsequent error-prone repair are important mechanisms in the formation of CNVs and pathogenic CNCs in humans.
Figures
Figure 1
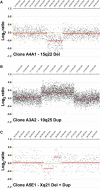
Examples of APH-Induced CNCs Detected by aCGH Representative data illustrating (A) a deletion at 15q22, (B) a duplication at 10q25, and (C) a tandem deletion and duplication at Xq21.
Figure 2
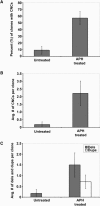
APH-Induced Replication Stress Creates Submicroscopic CNCs in Normal Human Fibroblasts (A) Percent of untreated (n = 11) and APH-treated (n = 14) clones containing one or more CNCs. (B) Average number of CNCs per clone in untreated and APH-treated clones. (C) Mean number of deletions (gray bars) and duplications (white bars) in untreated and APH-treated clones. Error bars indicate standard error.
Figure 3
CNCs Identified in APH-Treated Human Fibroblast Clones CNCs are mapped onto chromosome ideograms. Red bars to the left of a chromosome indicate deletions. Blue bars to the right of a chromosome indicate duplications.
Figure 4
Mapping of CNCs that Colocalize within the Same Chromosomal Bands or with Bands Containing Common Fragile Sites (A) Deletions (red bars) and duplications (blue bars) from chr. 3 hybrid clones and human fibroblast clones mapping to the same chromosomal bands. Asterisks (∗) indicate CNCs whose breakpoints were sequenced. (B) Examples of deletions (red bars) from human fibroblast clones mapping to chromosomal bands containing common fragile sites (black bars). Asterisks indicate CNCs whose breakpoints were sequenced.
Figure 5
PCR Strategy for Amplifying and Sequencing Deletion and Duplication Breakpoints Arrows represent PCR primers, designated P1 and P2, designed to flank deletion (A) and duplication (B) breakpoint junctions. Primers are designed to amplify breakpoints of duplications in any orientation.
Figure 6
Example of a Complex Deletion at 3q26.31 Detected in Chr. 3 Hybrid Clone 6-30 The red line indicates a deletion predicted by SegMNT at a 5000 bp interval average. Arrows indicate the positions of PCR assays used to detect the presence (+) or absence (−) of human chromosome 3 sequences in the clone. Blue areas represent undeleted regions, and pink areas represent deleted regions.
Similar articles
- Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture.
Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, de Vries BB, Gilissen C, Hoischen A, Nesteruk M, Wisniowiecka-Kowalnik B, Smyk M, Brunner HG, Cheung SW, van Kessel AG, Veltman JA, Stankiewicz P. Vissers LE, et al. Hum Mol Genet. 2009 Oct 1;18(19):3579-93. doi: 10.1093/hmg/ddp306. Epub 2009 Jul 3. Hum Mol Genet. 2009. PMID: 19578123 - Hydroxyurea induces de novo copy number variants in human cells.
Arlt MF, Ozdemir AC, Birkeland SR, Wilson TE, Glover TW. Arlt MF, et al. Proc Natl Acad Sci U S A. 2011 Oct 18;108(42):17360-5. doi: 10.1073/pnas.1109272108. Epub 2011 Oct 10. Proc Natl Acad Sci U S A. 2011. PMID: 21987784 Free PMC article. - De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining.
Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. Arlt MF, et al. PLoS Genet. 2012 Sep;8(9):e1002981. doi: 10.1371/journal.pgen.1002981. Epub 2012 Sep 20. PLoS Genet. 2012. PMID: 23028374 Free PMC article. - Origins and breakpoint analyses of copy number variations: up close and personal.
van Binsbergen E. van Binsbergen E. Cytogenet Genome Res. 2011;135(3-4):271-6. doi: 10.1159/000330267. Epub 2011 Aug 12. Cytogenet Genome Res. 2011. PMID: 21846967 Review.
Cited by
- Replication stress induces POLQ-mediated structural variant formation throughout common fragile sites after entry into mitosis.
Wilson TE, Ahmed S, Winningham A, Glover TW. Wilson TE, et al. Nat Commun. 2024 Nov 6;15(1):9582. doi: 10.1038/s41467-024-53917-8. Nat Commun. 2024. PMID: 39505880 Free PMC article. - FANCD2 genome binding is nonrandom and is enriched at large transcriptionally active neural genes prone to copy number variation.
Blaize JL, Garzon JLN, Howlett NG. Blaize JL, et al. Funct Integr Genomics. 2024 Oct 4;24(5):180. doi: 10.1007/s10142-024-01453-5. Funct Integr Genomics. 2024. PMID: 39365306 Free PMC article. - Proteogenomic characterization of skull-base chordoma.
Zhang Q, Xu Z, Han R, Wang Y, Ye Z, Zhu J, Cai Y, Zhang F, Zhao J, Yao B, Qin Z, Qiao N, Huang R, Feng J, Wang Y, Rui W, He F, Zhao Y, Ding C. Zhang Q, et al. Nat Commun. 2024 Sep 27;15(1):8338. doi: 10.1038/s41467-024-52285-7. Nat Commun. 2024. PMID: 39333076 Free PMC article. - Methods of Detection and Mechanisms of Origin of Complex Structural Genome Variations.
Poot M. Poot M. Methods Mol Biol. 2024;2825:39-65. doi: 10.1007/978-1-0716-3946-7_2. Methods Mol Biol. 2024. PMID: 38913302 Review. - Small polymorphisms are a source of ancestral bias in structural variant breakpoint placement.
Audano PA, Beck CR. Audano PA, et al. Genome Res. 2024 Feb 7;34(1):7-19. doi: 10.1101/gr.278203.123. Genome Res. 2024. PMID: 38176712 Free PMC article.
References
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. - PubMed
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Måner S., Massa H., Walker M., Chi M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
- Freeman J.L., Perry G.H., Feuk L., Redon R., McCarroll S.A., Altshuler D.M., Aburatani H., Jones K.W., Tyler-Smith C., Hurles M.E. Copy number variation: new insights in genome diversity. Genome Res. 2006;16:949–961. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources