Oncogenic gene fusions in epithelial carcinomas - PubMed (original) (raw)
Review
Oncogenic gene fusions in epithelial carcinomas
John R Prensner et al. Curr Opin Genet Dev. 2009 Feb.
Abstract
New discoveries regarding recurrent chromosomal aberrations in epithelial tumors have challenged the view that gene fusions play a minor role in these cancers. It is now known that recurrent fusions characterize significant subsets of prostate, breast, lung and renal-cell carcinomas, among others. This work has generated new insights into the molecular subtypes of tumors and highlighted important advances in bioinformatics, sequencing, and microarray technology as tools for gene fusion discovery. Given the ubiquity of tyrosine kinases and transcription factors in gene fusions, further interest in the potential 'druggability' of gene fusions with targeted therapeutics has also flourished. Nevertheless, the majority of chromosomal abnormalities in epithelial cancers remain uncharacterized, underscoring the limitations of our knowledge of carcinogenesis and the requirement for further research.
Figures
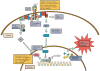
Figure 1. Biochemical Pathways in Gene Fusions
Biochemical effects of gene fusions cluster around tyrosine kinase (TK) signaling pathways, which alter the activity of intracellular proteins, and transcription factor (TF) activity, which control gene expression at the DNA level. Here we outline the examples of the Ras and PI-3K pathways, which are commonly involved downstream of TK activation and are frequently implicated in the oncogenic effects of gene fusions. PI-3K works via increased activity of the master regulator Akt, which controls many cellular processes including the nuclear TF NfκB. Likewise, the Ras-Raf-Mek-Erk pathway promotes activation of TFs, including Elk-1, which is a target of Erk. These signaling pathways and gene expression signatures result in the phenotypic qualities, such as invasiveness and increased proliferation, observed in cancers.
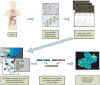
Figure 2. Gene Fusion Discovery and Targeted Therapy
Bioinformatic, sequencing and microarray methods are powerful tools for identifying potential gene fusions in epithelial cancers. By determining the genomic and transcriptomic events in human cancers, clinical management of the disease may be impacted, and gene fusions, such as the TMPRSS2-Ets fusions in prostate cancer, may serve as prominent therapeutic targets. If targeted therapeutics are successfully developed for critical oncogenes, clinical management of cancer may one day be determined based upon genetic evaluation of patient tumors.
Similar articles
- Morphologic features of carcinomas with recurrent gene fusions.
Qi M, Li Y, Liu J, Yang X, Wang L, Zhou Z, Han B. Qi M, et al. Adv Anat Pathol. 2012 Nov;19(6):417-24. doi: 10.1097/PAP.0b013e318273baae. Adv Anat Pathol. 2012. PMID: 23060067 Review. - Commonality but diversity in cancer gene fusions.
Rabbitts TH. Rabbitts TH. Cell. 2009 May 1;137(3):391-5. doi: 10.1016/j.cell.2009.04.034. Cell. 2009. PMID: 19410533 Review. - Evidence of recurrent gene fusions in common epithelial tumors.
Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Kumar-Sinha C, et al. Trends Mol Med. 2006 Nov;12(11):529-36. doi: 10.1016/j.molmed.2006.09.005. Epub 2006 Sep 29. Trends Mol Med. 2006. PMID: 17011825 Review. - Recurrent gene fusions in prostate cancer: their clinical implications and uses.
Hessels D, Schalken JA. Hessels D, et al. Curr Urol Rep. 2013 Jun;14(3):214-22. doi: 10.1007/s11934-013-0321-1. Curr Urol Rep. 2013. PMID: 23625457 Review.
Cited by
- Targeted next generation sequencing identifies somatic mutations and gene fusions in papillary thyroid carcinoma.
Lu Z, Zhang Y, Feng D, Sheng J, Yang W, Liu B. Lu Z, et al. Oncotarget. 2017 Jul 11;8(28):45784-45792. doi: 10.18632/oncotarget.17412. Oncotarget. 2017. PMID: 28507274 Free PMC article. - A Comprehensive Review and Androgen Deprivation Therapy and Its Impact on Alzheimer's Disease Risk in Older Men with Prostate Cancer.
Singh M, Agarwal V, Pancham P, Jindal D, Agarwal S, Rai SN, Singh SK, Gupta V. Singh M, et al. Degener Neurol Neuromuscul Dis. 2024 May 17;14:33-46. doi: 10.2147/DNND.S445130. eCollection 2024. Degener Neurol Neuromuscul Dis. 2024. PMID: 38774717 Free PMC article. Review. - Recent Advances in Metabolic Profiling And Imaging of Prostate Cancer.
Thapar R, Titus MA. Thapar R, et al. Curr Metabolomics. 2014 Apr;2(1):53-69. doi: 10.2174/2213235X02666140301002510. Curr Metabolomics. 2014. PMID: 25632377 Free PMC article. - IGF2/IGF1R Signaling as a Therapeutic Target in MYB-Positive Adenoid Cystic Carcinomas and Other Fusion Gene-Driven Tumors.
Andersson MK, Åman P, Stenman G. Andersson MK, et al. Cells. 2019 Aug 16;8(8):913. doi: 10.3390/cells8080913. Cells. 2019. PMID: 31426421 Free PMC article. Review. - Gene Duplication and Gene Fusion Are Important Drivers of Tumourigenesis during Cancer Evolution.
Glenfield C, Innan H. Glenfield C, et al. Genes (Basel). 2021 Aug 31;12(9):1376. doi: 10.3390/genes12091376. Genes (Basel). 2021. PMID: 34573358 Free PMC article. Review.
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
- Mitelman F, J B, Mertens F. Mitelman Database of Chromosome Aberrations in Cancer [online] 2008 Edited by.
- Heim S, Mitelman F. Molecular screening for new fusion genes in cancer. Nat Genet. 2008;40:685–686. - PubMed
- Mitelman F, Mertens F, Johansson B. Prevalence estimates of recurrent balanced cytogenetic aberrations and gene fusions in unselected patients with neoplastic disorders. Genes Chromosomes Cancer. 2005;43:350–366.. The most exhaustive study of chromosomal aberrations across cancer types The authors catalogue and analyze an immense amount of data to distill the most accurate account of chromosomal abnormalities in cancer available.
Publication types
MeSH terms
Substances
Grants and funding
- P50 CA069568/CA/NCI NIH HHS/United States
- R01 CA132874/CA/NCI NIH HHS/United States
- U01 CA111275/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical