Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites - PubMed (original) (raw)
Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites
William R Wikoff et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Although it has long been recognized that the enteric community of bacteria that inhabit the human distal intestinal track broadly impacts human health, the biochemical details that underlie these effects remain largely undefined. Here, we report a broad MS-based metabolomics study that demonstrates a surprisingly large effect of the gut "microbiome" on mammalian blood metabolites. Plasma extracts from germ-free mice were compared with samples from conventional (conv) animals by using various MS-based methods. Hundreds of features were detected in only 1 sample set, with the majority of these being unique to the conv animals, whereas approximately 10% of all features observed in both sample sets showed significant changes in their relative signal intensity. Amino acid metabolites were particularly affected. For example, the bacterial-mediated production of bioactive indole-containing metabolites derived from tryptophan such as indoxyl sulfate and the antioxidant indole-3-propionic acid (IPA) was impacted. Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium Clostridium sporogenes. Multiple organic acids containing phenyl groups were also greatly increased in the presence of gut microbes. A broad, drug-like phase II metabolic response of the host to metabolites generated by the microbiome was observed, suggesting that the gut microflora has a direct impact on the drug metabolism capacity of the host. Together, these results suggest a significant interplay between bacterial and mammalian metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
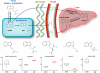
The microbione exerts a broad effect on mammalian biochemistry. (Upper) The experimental workflow for the metabolomics studies performed. (Lower) Major differences are observed in the profiles of GF and conv plasma obtained employing + ESI. PCA analysis of the profiling data from XCMS (Left) shows an excellent separation between sample groups. As shown in the Venn diagram (Right), most species are observed in both GF and conv samples, but a significant number are unique to a given sample set, with ≈3-fold more ions unique to the conv sample.
Fig. 2.
The diversity of indole-containing compounds in serum is affected by the microbiome. (Upper) Various indole-containing molecules arise only in the presence of the microbiome and ultimately enter the plasma by means of different routes. (Lower) Differences in the plasma levels of indole-containing compounds attributed to the action of the microbiome. The integrated signal intensities plotted on the y axes are reduced by a factor of 1,000.
Fig. 3.
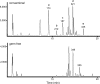
Sulfate profiling based on constant neutral loss scanning of 80 m/z in − ESI mode for conv (Upper) and GF (Lower) pooled plasma samples. The m/z values for several species are listed above their respective peak. Identified sulfates include: phenyl sulfate (a), indoxyl sulfate (b), _p_-cresol sulfate (c), equol sulfate (d), and methyl equol sulfate (e).
Fig. 4.
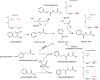
Differences in the plasma levels of various glycine-conjugated compounds attributed to the action of the microbiome. Potential metabolic pathways leading to the formation of hippuric acid as well as 3 other glycine conjugates arising from the presence of the microbiome are provided. The integrated signal intensities plotted on the y axes are reduced by a factor of 1,000.
Similar articles
- Metabolomics approaches for characterizing metabolic interactions between host and its commensal microbes.
Xie G, Zhang S, Zheng X, Jia W. Xie G, et al. Electrophoresis. 2013 Oct;34(19):2787-98. doi: 10.1002/elps.201300017. Epub 2013 Aug 16. Electrophoresis. 2013. PMID: 23775228 Review. - Tissue-wide metabolomics reveals wide impact of gut microbiota on mice metabolite composition.
Zarei I, Koistinen VM, Kokla M, Klåvus A, Babu AF, Lehtonen M, Auriola S, Hanhineva K. Zarei I, et al. Sci Rep. 2022 Sep 2;12(1):15018. doi: 10.1038/s41598-022-19327-w. Sci Rep. 2022. PMID: 36056162 Free PMC article. - Evaluating effects of penicillin treatment on the metabolome of rats.
Sun J, Schnackenberg LK, Khare S, Yang X, Greenhaw J, Salminen W, Mendrick DL, Beger RD. Sun J, et al. J Chromatogr B Analyt Technol Biomed Life Sci. 2013 Aug 1;932:134-43. doi: 10.1016/j.jchromb.2013.05.030. Epub 2013 Jun 7. J Chromatogr B Analyt Technol Biomed Life Sci. 2013. PMID: 23831706 - Disorder of gut amino acids metabolism during CKD progression is related with gut microbiota dysbiosis and metagenome change.
Liu Y, Li J, Yu J, Wang Y, Lu J, Shang EX, Zhu Z, Guo J, Duan J. Liu Y, et al. J Pharm Biomed Anal. 2018 Feb 5;149:425-435. doi: 10.1016/j.jpba.2017.11.040. Epub 2017 Nov 15. J Pharm Biomed Anal. 2018. PMID: 29169110 - Indolic Structure Metabolites as Potential Biomarkers of Non-infectious Diseases.
Beloborodova NV, Chernevskaya EA, Getsina ML. Beloborodova NV, et al. Curr Pharm Des. 2021;27(2):238-249. doi: 10.2174/1381612826666201022121653. Curr Pharm Des. 2021. PMID: 33092503 Review.
Cited by
- Delving the depths of 'terra incognita' in the human intestine - the small intestinal microbiota.
Yilmaz B, Macpherson AJ. Yilmaz B, et al. Nat Rev Gastroenterol Hepatol. 2025 Jan;22(1):71-81. doi: 10.1038/s41575-024-01000-4. Epub 2024 Oct 23. Nat Rev Gastroenterol Hepatol. 2025. PMID: 39443711 Review. - Characterizing the influence of gut microbiota on host tryptophan metabolism with germ-free pigs.
Liu B, Yu D, Sun J, Wu X, Xin Z, Deng B, Fan L, Fu J, Ge L, Ren W. Liu B, et al. Anim Nutr. 2022 Jul 31;11:190-200. doi: 10.1016/j.aninu.2022.07.005. eCollection 2022 Dec. Anim Nutr. 2022. PMID: 36263410 Free PMC article. - Metabolomics and cardiovascular biomarker discovery.
Rhee EP, Gerszten RE. Rhee EP, et al. Clin Chem. 2012 Jan;58(1):139-47. doi: 10.1373/clinchem.2011.169573. Epub 2011 Nov 22. Clin Chem. 2012. PMID: 22110018 Free PMC article. Review. - Emerging Roles of Gut Serotonin in Regulation of Immune Response, Microbiota Composition and Intestinal Inflammation.
Grondin JA, Khan WI. Grondin JA, et al. J Can Assoc Gastroenterol. 2023 Jul 13;7(1):88-96. doi: 10.1093/jcag/gwad020. eCollection 2024 Feb. J Can Assoc Gastroenterol. 2023. PMID: 38314177 Free PMC article. Review. - Factors Affecting Gut Microbiome in Daily Diet.
Su Q, Liu Q. Su Q, et al. Front Nutr. 2021 May 10;8:644138. doi: 10.3389/fnut.2021.644138. eCollection 2021. Front Nutr. 2021. PMID: 34041257 Free PMC article. Review.
References
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. - PubMed
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical