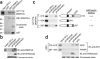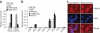PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing - PubMed (original) (raw)
. 2009 Mar;16(3):304-311.
doi: 10.1038/nsmb.1568. Epub 2009 Feb 22.
Gerhard Rank # 1, Yuen T Tan 1, Haitao Li 3, Robert L Moritz 4, Richard J Simpson 4, Loretta Cerruti 1, David J Curtis 1, Dinshaw J Patel 3, C David Allis 5, John M Cunningham 6, Stephen M Jane 1 7
Affiliations
- PMID: 19234465
- PMCID: PMC5120857
- DOI: 10.1038/nsmb.1568
PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing
Quan Zhao et al. Nat Struct Mol Biol. 2009 Mar.
Abstract
Mammalian gene silencing is established through methylation of histones and DNA, although the order in which these modifications occur remains contentious. Using the human beta-globin locus as a model, we demonstrate that symmetric methylation of histone H4 arginine 3 (H4R3me2s) by the protein arginine methyltransferase PRMT5 is required for subsequent DNA methylation. H4R3me2s serves as a direct binding target for the DNA methyltransferase DNMT3A, which interacts through the ADD domain containing the PHD motif. Loss of the H4R3me2s mark through short hairpin RNA-mediated knockdown of PRMT5 leads to reduced DNMT3A binding, loss of DNA methylation and gene activation. In primary erythroid progenitors from adult bone marrow, H4R3me2s marks the inactive methylated globin genes coincident with localization of PRMT5. Our findings define DNMT3A as both a reader and a writer of repressive epigenetic marks, thereby directly linking histone and DNA methylation in gene silencing.
Figures
Figure 1
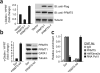
PRMT5 symmetrically dimethylates histone H4R3 on the gene encoding γ-globin. (a) SimplyBlue Safestain of an SDS-PAGE gel of α-Flag antibody immunoprecipitates from K562 cells transfected with NF-E4-Flag, or vector alone (control) before analysis by mass spectrometry. The bands corresponding to PRMT5, NF-E4, polyubiquitinated NF-E4 (ref. 52) and the known PRMT5 partner proteins nucleolin and pICln are shown. Hash marks indicate bands that correspond to common background proteins including keratin, tubulins and ribosomal proteins. Asterisks indicate immunoglobulin chains. The PRMT5 peptide sequences identified are shown below. (b) Co-immunoprecipitation of endogenous PRMT5 and NF-E4 from K562 cells. (c) Interaction of the endogenous PRMT5 and NF-E4 with the γ-promoter by ChIP assays. (d) In vitro methyltransferase assays of Flag immunoprecipitates from K562 cells expressing PRMT5-f and K562 cells expressing PRMT5Δ-f. Autoradiographs (upper panels) and Coomassiestained gels (lower panels) are shown for each. (e) The HMTase assays with free histones detailed in d were immunoprecipitated with antibodies to histone H4R3me2s or normal rabbit IgG, and the radioactivity was quantitated in the respective precipitates. CPM, counts per minute. (f) H4R3me2s enrichment at the γ-promoter was measured by ChIP in K562 cells expressing PRMT5-f or PRMT5Δ-f. Error bars show s.d.
Figure 2
PRMT5 mediates transcriptional silencing of the γ-gene. (a) Extracts from PRMT5-f, PRMT5Δ-f or vector control K562 cells were analyzed by western blot with anti-Flag or PRMT5 antibodies (right panel). RNA from these cells was analyzed by Q-RT-PCR with primers specific for the gene encoding γ-globin, and the signals were normalized against HPRT mRNA levels (left panel). (b) K562 cells expressing either an shRNA to PRMT5 (PRMT5-kd) or a scrambled sequence (Scr) were analyzed by western blot (with antibodies indicated, right panel) and Q-RT-PCR (left panel) as ina. Error bars show s.d. (c) H4R3me2s enrichment at the γ-promoter was measured by ChIP in PRMT5-kd and Scr cells. Error bars show s.d.
Figure 3
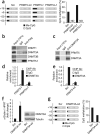
PRMT5 and DNMT3A function cooperatively in gene silencing. (a) Effect of perturbed PRMT5 expression on DNA methylation at the human γ-gene. Each row shows the methylation status of individual CpG dinucleotides derived from sequence analysis of at least 40 individual cloned PCR products of the γ-genes following bisulfite modification from PRMT5-f, PRMT5Δ-f, PRMT-kd and the scrambled control (Scr) K562 cells. The differences between the three lines and the Scr were highly significant (P< o 0.01). The numbers on the left represent the positions of the CpG dinucleotides relative to the transcriptional start site of the γ-gene. The results are quantitated in the bar graph. ND, not detectable. (b) DNMT3A, but not DNMT1 or DNMT3B, co-immunoprecipitates with PRMT5-f from K562 cells. High salt extraction (420 mM NaCl) was used for the cellular extract preparation. (c) Co-immunoprecipitation of endogenous DNMT3A and PRMT5 from K562 cells. High salt extraction (420 mM NaCl) was used for the cellular extract preparation. (d) DNMT3A binding at the γ-promoter was measured by ChIP in PRMT5-f and PRMT5Δ-f cells. Error bars show s.d. (e) DNMT3A binding at the γ-promoter was measured by ChIP in PRMT-kd and Scr cells as above. (f) K562 cells expressing either an shRNA to DNMT3A (DNMT3A-kd) or a scrambled sequence (Scr) were analyzed by western blot, with indicated antibodies (right panel), and Q-RT-PCR with primers specific for the γ-globin genes, with the signal normalized against HPRT mRNA levels (left panel). (g) Effect of DNMT3A knockdown on DNA methylation at the human γ-genes as detailed in a. The difference between the two lines was significant (P < 0.02).
Figure 4
DNMT3A binds specifically to histone H4 carrying the R3me2s modification. (a) Binding of 35S-labeled _in vitro_transcribed and translated (IVTT) DNMT3A to purified GST, GST-PRMT5 and GSTPRMT5D. Top panel, autoradiograph; bottom panel, Coomassie. Input represents 30% of the in vitro translated DNMT3A used in the assay. (b) Binding of DNMT3A to N-terminal peptides of histone H4 with the Arg3 residue unmethylated, symmetrically methylated or asymmetrically methylated. Specifically bound protein was visualized by western blot with anti-DNMT3A antibody after SDS-PAGE. Input represents 10% of the cellular extract used in the assay. The H4R3me2s modification of the synthesized peptide was confirmed by immunoblot. Coomassie staining shows equivalent loading of the three peptides on a 20% (w/v) SDS-PAGE gel. (c) Peptide pulldown assays as described in b, with 35S-labeled fragments of DNMT3A as shown in the schematic. Numbers refer to amino acids. The 1–354 construct contains the PWWP module but lacks its adjacent C-terminal helical motif. The 1–587 construct contains the GATA and PHD domains of ADD, but lacks an adjacent C-terminal helix. (d) Peptide pulldown assays as described in b, with purified GST, GST-DNMT3A (281–424, containing the PWWP domain) and GST-DNMT3A (479–610, containing the ADD domain). Specifically bound protein was visualized by western blot with anti-GST antibody after SDS-PAGE. Input represents 5% of the GST fusion proteins used in the assay. The H4R3me2s modification of the synthesized peptide was confirmed by immunoblot. Coomassie staining shows equivalent loading of the four peptides.
Figure 5
Role of PRMT5 in developmental globin gene silencing. (a) H4R3me2s and RNA polymerase II enrichment at the γ-promoter was measured by ChIP in chromatin fractions from erythroid progenitors from cord blood and adult bone marrow. (b) Localization of PRMT5, NF-E4 and H4R3me2s across the β-globin locus measured by ChIP in chromatin fractions from erythroid progenitors from adult bone marrow. The precipitated DNA was amplified with primers specific for the indicated regions of the β-globin locus. HS, hypersensitive site; Pro, promoter; G/Aγ, intergenic region between Gγ-globin and Aγ-globin genes. Error bars show s.d. (c) Cellular localization of PRMT5 in erythroid progenitors from cord blood and adult bone marrow shown by immunofluorescence with anti-PRMT5 antibody and DAPI nuclear counterstaining. Scale bar, 10 μm.
Figure 6
Model of PRMT5-induced silencing of gene expression. Symmetric dimethylation of H4R3 by PRMT5 induces direct binding of DNMT3A, resulting in methylation of adjacent CpG dinucleotides and gene silencing.
Similar articles
- Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression.
Rank G, Cerruti L, Simpson RJ, Moritz RL, Jane SM, Zhao Q. Rank G, et al. Blood. 2010 Sep 2;116(9):1585-92. doi: 10.1182/blood-2009-10-251116. Epub 2010 May 21. Blood. 2010. PMID: 20495075 Free PMC article. - PRMT5-mediated histone H4 arginine-3 symmetrical dimethylation marks chromatin at G + C-rich regions of the mouse genome.
Girardot M, Hirasawa R, Kacem S, Fritsch L, Pontis J, Kota SK, Filipponi D, Fabbrizio E, Sardet C, Lohmann F, Kadam S, Ait-Si-Ali S, Feil R. Girardot M, et al. Nucleic Acids Res. 2014 Jan;42(1):235-48. doi: 10.1093/nar/gkt884. Epub 2013 Oct 3. Nucleic Acids Res. 2014. PMID: 24097435 Free PMC article. - The role of WDR5 in silencing human fetal globin gene expression.
Xu Z, He Y, Ju J, Rank G, Cerruti L, Ma C, Simpson RJ, Moritz RL, Jane SM, Zhao Q. Xu Z, et al. Haematologica. 2012 Nov;97(11):1632-40. doi: 10.3324/haematol.2012.061937. Epub 2012 Jun 11. Haematologica. 2012. PMID: 22689669 Free PMC article. - Molecular coupling of DNA methylation and histone methylation.
Hashimoto H, Vertino PM, Cheng X. Hashimoto H, et al. Epigenomics. 2010 Oct;2(5):657-69. doi: 10.2217/epi.10.44. Epigenomics. 2010. PMID: 21339843 Free PMC article. Review. - Versatility of PRMT5-induced methylation in growth control and development.
Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S. Karkhanis V, et al. Trends Biochem Sci. 2011 Dec;36(12):633-41. doi: 10.1016/j.tibs.2011.09.001. Epub 2011 Oct 3. Trends Biochem Sci. 2011. PMID: 21975038 Free PMC article. Review.
Cited by
- High-throughput development and characterization of new functional nanobodies for gene regulation and epigenetic control in human cells.
Wan J, Thurm AR, Allen SJ, Ludwig CH, Patel AN, Bintu L. Wan J, et al. bioRxiv [Preprint]. 2024 Nov 3:2024.11.01.621523. doi: 10.1101/2024.11.01.621523. bioRxiv. 2024. PMID: 39554150 Free PMC article. Preprint. - Deciphering the potential role of post-translational modifications of histones in gastrointestinal cancers: a proteomics-based review with therapeutic challenges and opportunities.
Farrokhi Yekta R, Farahani M, Koushki M, Amiri-Dashatan N. Farrokhi Yekta R, et al. Front Oncol. 2024 Oct 21;14:1481426. doi: 10.3389/fonc.2024.1481426. eCollection 2024. Front Oncol. 2024. PMID: 39497715 Free PMC article. Review. - Emerging roles of cancer-associated histone mutations in genomic instabilities.
Yadav P, Jain R, Yadav RK. Yadav P, et al. Front Cell Dev Biol. 2024 Oct 8;12:1455572. doi: 10.3389/fcell.2024.1455572. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39439908 Free PMC article. Review. - Dysregulation of epigenetic modifications in inborn errors of immunity.
Xiao Z, He R, Zhao Z, Chen T, Ying Z. Xiao Z, et al. Epigenomics. 2024;16(19-20):1301-1313. doi: 10.1080/17501911.2024.2410695. Epub 2024 Oct 15. Epigenomics. 2024. PMID: 39404224 Review. - CircGSK3β mediates PD-L1 transcription through miR-338-3p/PRMT5/H3K4me3 to promote breast cancer cell immune evasion and tumor progression.
Liang L, Gao M, Li W, Tang J, He Q, Zeng F, Cao J, Liu S, Chen Y, Li X, Zhou Y. Liang L, et al. Cell Death Discov. 2024 Oct 4;10(1):426. doi: 10.1038/s41420-024-02197-8. Cell Death Discov. 2024. PMID: 39366935 Free PMC article.
References
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. - PubMed
- Bird AP, Wolffe AP. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99:451–454. - PubMed
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM040922/GM/NIGMS NIH HHS/United States
- P01 HL53749-03/HL/NHLBI NIH HHS/United States
- R01 HL069232/HL/NHLBI NIH HHS/United States
- P01 HL053749/HL/NHLBI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 HL69232-01/HL/NHLBI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases