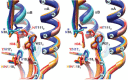Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses - PubMed (original) (raw)
doi: 10.1038/nsmb.1566. Epub 2009 Feb 22.
William C Hwang, Sandra Perez, Ge Wei, Daniel Aird, Li-mei Chen, Eugenio Santelli, Boguslaw Stec, Greg Cadwell, Maryam Ali, Hongquan Wan, Akikazu Murakami, Anuradha Yammanuru, Thomas Han, Nancy J Cox, Laurie A Bankston, Ruben O Donis, Robert C Liddington, Wayne A Marasco
Affiliations
- PMID: 19234466
- PMCID: PMC2692245
- DOI: 10.1038/nsmb.1566
Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses
Jianhua Sui et al. Nat Struct Mol Biol. 2009 Mar.
Abstract
Influenza virus remains a serious health threat, owing to its ability to evade immune surveillance through rapid genetic drift and reassortment. Here we used a human non-immune antibody phage-display library and the H5 hemagglutinin ectodomain to select ten neutralizing antibodies (nAbs) that were effective against all group 1 influenza viruses tested, including H5N1 'bird flu' and the H1N1 'Spanish flu'. The crystal structure of one such nAb bound to H5 shows that it blocks infection by inserting its heavy chain into a conserved pocket in the stem region, thus preventing membrane fusion. Nine of the nAbs employ the germline gene VH1-69, and all seem to use the same neutralizing mechanism. Our data further suggest that this region is recalcitrant to neutralization escape and that nAb-based immunotherapy is a promising strategy for broad-spectrum protection against seasonal and pandemic influenza viruses.
Figures
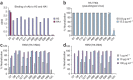
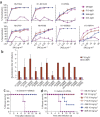
Figure 1. In vitro binding and neutralization of anti-H5 antibodies.
(a) The ten antibodies were converted to soluble scFv-Fcs (scFv linked to the hinge, CH2 and CH3 domains of human IgG1) and evaluated for binding to trimeric H5-TH04 or monomeric HA1 of H5-TH04 coated on an ELISA plate. The H5 scFv-Fcs recognize trimeric H5 but not HA1. An antibody raised against HA1 (2A) recognized both. (b) Neutralization of H5-TH04–pseudotyped viruses (virus-like particles with HIV-1 only cores that display H5 on their surface). Percentage of neutralization at two concentrations is shown with s.d. The mAb 80R was used as a negative control (Ctrl.). (c,d) Neutralization of wild-type H5-VN04 and H5-IN05 by the ten scFv-Fcs at three concentrations using a plaque reduction assay. Results are consistent with those obtained from a microneutralization assay (data not shown).
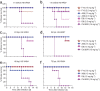
Figure 2. Prophylactic and therapeutic efficacy of anti-H5 nAbs in mice.
(a,b) Prophylactic efficacy. Percentage of survival of mice treated with anti-H5 nAbs or control mAb 1 h before lethal challenge by intranasal inoculation with H5-VN04 (a) or H5-HK97 (b) viruses. (c–f). Therapeutic efficacy. Mice were inoculated with H5-VN04 and injected with nAbs at 24 h, 48 h of 72 hpi (c,e,f) or with H5-HK97 at 24 hpi (d).
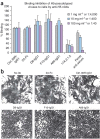
Figure 3. Neutralization mechanism.
(a) nAbs do not inhibit cell binding of full-length hemagglutinin from H5-TH04–pseudotyped HIV-1 viruses. None of the three nAb-treated viruses inhibited cell binding. Mouse anti-H5 mAb, 17A2.1.2 and ferret anti-H5N1 serum, which inhibit hemagglutination, were used as positive controls. Anti-SARS spike protein (80R) and anti-HA1 (2A) were used as negative controls. Error bars represent s.d. (b) All three nAbs inhibit cell fusion. HeLa cells were transfected with H5-TH04–expressing plasmid and exposed to a pH 5.0 buffer for 4 min in the presence or absence of nAbs. Syncytia formation induced by the brief exposure to pH 5.0 was completely inhibited by D8, F10 and A66, at 20 μg ml−1 (∼0.13 μM), whereas controls (80R and anti-HA1 mAb (2A) at the same concentration had no effect.
Figure 4. Structure of the H5–F10 complex.
(a) Structure of the H5 trimer bound to F10 (scFv). H5 is similar to the uncomplexed structure (pairwise r.m.s. deviation (Cα) = 1.0 and 0.63 Å for two independent trimers). HA1, HA2, the αA helix of HA2, the fusion peptide (FP) and F10 (VH and VL) are color coded. The third F10 molecule is hidden behind the stem. (b) Close-up of the epitope showing H5 as a molecular surface, with selected epitope residues labeled. The fusion peptide is in green. The tip of F10 (red ribbon) and selected CDR side chains are shown. Of 1,500 Å2 buried surface at the interface, 43% involves hydrophobic interactions. (c) Surface of the central stem region, showing two H5 monomers. One monomer has HA1 (yellow) and HA2 (blue) colored differently; the path of the FP through the epitope (red) is outlined, and mutations that do not affect binding are colored cyan (Fig. 4d). The fusion peptides (FP and FP′) are labeled in both monomers. Epitope residues are labeled white (HA2) or yellow (HA1), and the position of buried residue H1112 is shown as a black ball labeled 'H'. (d) Binding of the three nAbs to H5 mutants in the αA helix, transiently transfected into 293T cells. Note the similar response to all mutants tested. Mutations were made either to alanine or to the corresponding H7 residue; 24 h after transfection, nAbs or ferret anti-H5N1 serum was used to stain the transfected cells. Fluorescent intensity was normalized against ferret anti-serum (100%) to account for different expression levels.
Figure 5. Sequence conservation in hemagglutinin groups, clusters and subtypes at the F10 epitope.
Circles below residue numbers indicate estimated contribution to the binding energy at each position: red, strong; yellow, intermediate; blue, neutral. Residues without a circle are not directly involved in the epitope but are discussed in the text. Colored highlighting on the sequences indicates conservation within clusters and groups, with orange indicating high conservation or invariance. Other colors (for example, yellow, cyan and pink) highlight residues that are cluster or subtype specific. The network of interhelical contacts that stabilize the fusogenic structure are indicated below the HA2 sequences. Subtypes that can be recognized/neutralized by F10 are indicated with '+' on the far right. (+) or (−) indicates a predicted positive or negative binding, respectively.
Figure 6. Cross-subtype neutralization by nAbs.
(a) nAbs D8, F10 and A66 all neutralized indicated pseudotyped viruses (strains described below). Error bars indicate s.d. (b) Microneutralization assay. Neutralization titers (0.1 mg ml−1 antibody stock solution) of nAb F10 against wild-type H5N1, H1N1, H2N2, H6N1, H6N2, H8N4, H9N2 and H3N2 virus strains. 80R is the negative control. Vertical bars and whiskers represent the lowest and the highest neutralization titer (2χ, values of χ are shown on the y axis), respectively, of two or three independent experiments. (c,d) Prophylactic efficacy against two H1N1 strains in mice. Percentage of survival of mice treated with anti-H5 nAbs or control mAb are shown before lethal challenge by intranasal inoculation with H1-WSN33 (c) or H1-PR34 (d) viruses. Complete viral strain designations are: H1-OH83 (A/Ohio/83 (H1N1)); H1-PR34 (A/Puerto Rico/8/34 (H1N1)); H1-SC1918 (A/South Carolina/1/1918 (H1N1)); H1-WSN33 (A/WSN/1933 (H1N1)); H2-AA60 (A/Ann Arbor/6/60 (H2N2)); H2-JP57 (A/Japan/305/57(H2N2)); H3-SY97 (A/Sydney/5/97(H3N2)); H6-HK99 (A/quail/Hong Kong/1721-30/99(H6N1)); H6-NY98 (A/chicken/New York/14677-13/1998 (H6N2)); H7-FP34 (A/FPV/Rostock/34 (H7N1)); H8-ON68 (A/turkey/Ontario/6118/68); H9-HK(G9)97 (A/chicken/HongKong/G9/97 (H9N2)); H9-HK99 (A/HongKong/1073/99 (H9N2)); H11-MP74 (A/duck/Memphis/546/74 (H11N9)).
Figure 7. Three-dimensional comparison of the F10 epitope in group 1 and group 2 hemagglutinins.
Stereo overlay of crystal structures of the five known hemagglutinin subtypes in the region of the F10 epitope, showing conservation and differences between the two phylogenetic groups. H1, H5 and H9 (group 1) are in shades of red and yellow (PDB
1RU7
,
2IBX
and
1JSD
); H3 and H7 (group 2) are in shades of blue (PDB
1MQL
and
1TI8
). R.m.s. differences for pairwise overlays are 0.56 ± 0.11 Å (observed range, group 1), 0.75 Å (group 2) and 1.21 ± 0.12 Å between groups. Consistent differences between phylogenetic groups include the orientation of Trp212 and alternative side chain directions at 181 and 381, which are linked to the packing of buried His1112 (the putative pH trigger in group 1; absent in group 2), and the burial of the larger tyrosine (group 1) versus histidine (the putative pH trigger in group 2) at 171. Of particular note, Asn381 is glycosylated in four members of the group 2 clusters. Other epitope residues are indicated by numbered light blue circles.
Comment in
- Universal epitopes of influenza virus hemagglutinins?
Wang TT, Palese P. Wang TT, et al. Nat Struct Mol Biol. 2009 Mar;16(3):233-4. doi: 10.1038/nsmb.1574. Epub 2009 Feb 22. Nat Struct Mol Biol. 2009. PMID: 19234464 No abstract available.
Similar articles
- A non-VH1-69 heterosubtypic neutralizing human monoclonal antibody protects mice against H1N1 and H5N1 viruses.
De Marco D, Clementi N, Mancini N, Solforosi L, Moreno GJ, Sun X, Tumpey TM, Gubareva LV, Mishin V, Clementi M, Burioni R. De Marco D, et al. PLoS One. 2012;7(4):e34415. doi: 10.1371/journal.pone.0034415. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496802 Free PMC article. - Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells.
Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. Throsby M, et al. PLoS One. 2008;3(12):e3942. doi: 10.1371/journal.pone.0003942. Epub 2008 Dec 16. PLoS One. 2008. PMID: 19079604 Free PMC article. - Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
Lin Q, Li T, Chen Y, Lau SY, Wei M, Zhang Y, Zhang Z, Yao Q, Li J, Li Z, Wang D, Zheng Q, Yu H, Gu Y, Zhang J, Chen H, Li S, Xia N. Lin Q, et al. J Virol. 2018 Aug 16;92(17):e00547-18. doi: 10.1128/JVI.00547-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29925655 Free PMC article. - A novel humanized antibody neutralizes H5N1 influenza virus via two different mechanisms.
Tan Y, Ng Q, Jia Q, Kwang J, He F. Tan Y, et al. J Virol. 2015 Apr;89(7):3712-22. doi: 10.1128/JVI.03014-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609802 Free PMC article. - One step closer to universal influenza epitopes.
Li OT, Poon LL. Li OT, et al. Expert Rev Anti Infect Ther. 2009 Aug;7(6):687-90. doi: 10.1586/eri.09.48. Expert Rev Anti Infect Ther. 2009. PMID: 19681695 Review.
Cited by
- Towards a universal influenza vaccine: volunteer virus challenge studies in quarantine to speed the development and subsequent licensing.
Oxford JS. Oxford JS. Br J Clin Pharmacol. 2013 Aug;76(2):210-6. doi: 10.1111/bcp.12146. Br J Clin Pharmacol. 2013. PMID: 23617282 Free PMC article. Review. - Protection From Influenza by Intramuscular Gene Vector Delivery of a Broadly Neutralizing Nanobody Does Not Depend on Antibody Dependent Cellular Cytotoxicity.
Del Rosario JMM, Smith M, Zaki K, Risley P, Temperton N, Engelhardt OG, Collins M, Takeuchi Y, Hufton SE. Del Rosario JMM, et al. Front Immunol. 2020 May 29;11:627. doi: 10.3389/fimmu.2020.00627. eCollection 2020. Front Immunol. 2020. PMID: 32547534 Free PMC article. - A critical HA1 neutralizing domain of H5N1 influenza in an optimal conformation induces strong cross-protection.
Du L, Zhao G, Sun S, Zhang X, Zhou X, Guo Y, Li Y, Zhou Y, Jiang S. Du L, et al. PLoS One. 2013;8(1):e53568. doi: 10.1371/journal.pone.0053568. Epub 2013 Jan 8. PLoS One. 2013. PMID: 23320093 Free PMC article. - Heterosubtypic antibodies to influenza A virus have limited activity against cell-bound virus but are not impaired by strain-specific serum antibodies.
Wyrzucki A, Bianchi M, Kohler I, Steck M, Hangartner L. Wyrzucki A, et al. J Virol. 2015 Mar;89(6):3136-44. doi: 10.1128/JVI.03069-14. Epub 2014 Dec 31. J Virol. 2015. PMID: 25552718 Free PMC article. - Influenza virus glycoprotein-reactive human monoclonal antibodies.
Li Y, Wang L, Si H, Yu Z, Tian S, Xiang R, Deng X, Liang R, Jiang S, Yu F. Li Y, et al. Microbes Infect. 2020 Jul-Aug;22(6-7):263-271. doi: 10.1016/j.micinf.2020.06.003. Epub 2020 Jun 19. Microbes Infect. 2020. PMID: 32569735 Free PMC article. Review.
References
- WHO. Factsheet 211: influenza. World Health Organization 〈http://www.who.int/mediacentre/factsheets/2003/fs211/en/〉 (2003).
- WHO. Global influenza surveillance. World Health Organization 〈http://www.who.int/csr/disease/influenza/influenzanetwork/en/index.html〉 (2008).
Publication types
MeSH terms
Substances
Grants and funding
- P01-AI055789/AI/NIAID NIH HHS/United States
- U01 AI074518/AI/NIAID NIH HHS/United States
- U01-AI07518-01/AI/NIAID NIH HHS/United States
- P41 RR-01081/RR/NCRR NIH HHS/United States
- P01 AI055789/AI/NIAID NIH HHS/United States
- P01 AI055789-040001/AI/NIAID NIH HHS/United States
- P41 RR001081/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous