Mutations in the nonstructural protein 3A confer resistance to the novel enterovirus replication inhibitor TTP-8307 - PubMed (original) (raw)
doi: 10.1128/AAC.00934-08. Epub 2009 Feb 23.
Hendrik Jan Thibaut, Lonneke van der Linden, Kjerstin Lanke, Ward Heggermont, Stephen Ireland, Robert Andrews, Murty Arimilli, Taleb H Al-Tel, Erik De Clercq, Frank van Kuppeveld, Johan Neyts
Affiliations
- PMID: 19237651
- PMCID: PMC2681499
- DOI: 10.1128/AAC.00934-08
Mutations in the nonstructural protein 3A confer resistance to the novel enterovirus replication inhibitor TTP-8307
Armando M De Palma et al. Antimicrob Agents Chemother. 2009 May.
Abstract
A novel compound, TTP-8307, was identified as a potent inhibitor of the replication of several rhino- and enteroviruses. TTP-8307 inhibits viral RNA synthesis in a dose-dependent manner, without affecting polyprotein synthesis and/or processing. Drug-resistant variants of coxsackievirus B3 were all shown to carry at least one amino acid mutation in the nonstructural protein 3A. In particular, three mutations located in a nonstructured region preceding the hydrophobic domain (V45A, I54F, and H57Y) appeared to contribute to the drug-resistant phenotype. This region has previously been identified as a hot sport for mutations that resulted in resistance to enviroxime, the sole 3A-targeting enterovirus inhibitor reported thus far. This was corroborated by the fact that TTP-8307 and enviroxime proved cross-resistant. It is hypothesized that TTP-8307 and enviroxime disrupt proper interactions of 3A(B) with other viral or cellular proteins that are required for efficient replication.
Figures
FIG. 1.
Structural formulae of TTP-8307 (A) and enviroxime (B).
FIG. 2.
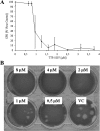
Dose-dependent inhibition of viral replication by TTP-8307. Vero cell cultures infected with CVB3 were treated with different concentrations of TTP-8307, and the effect on viral replication was monitored at day 3 postinfection. (A) Effect on virus-induced CPE formation (using an MTS-based cell protection assay). (B) Plaque reduction. The data represent averages ± the standard deviations (SD) from three independent experiments.
FIG. 3.
TTP-8307 inhibits accumulation of CVB3 viral RNA without affecting polyprotein processing. (A) Time of drug addition. TTP-8307 (25 μM) was added to CVB3-infected cell cultures at 1-h intervals, starting at 1 h before infection. At 8 h postinfection, the time needed for a single replication cycle, intra- and extracellular viral RNA was collected and quantified by means of real-time qRT-PCR. Values were standardized to the 0-h time point. The data represent averages ± the SD from two independent experiments. (B) Analysis of viral RNA accumulation with subgenomic replicon pCB53/T7-Luc. Accumulation of viral (+)RNA was monitored after transfection of BGM cells (in the presence or absence of TTP-8307) with RNA derived from a chimeric subgenomic replicon (pCB53/T7-Luc). At the indicated times posttransfection, the luciferase activity was quantified and expressed in (relative) light units [(R)LU]. The data represent averages ± the SD from three independent experiments. (C) Effect of TTP-8307 on polyprotein processing. BGM cells grown to confluence in 24-well plates were infected with CVB3. At 5h30 min postinfection, cells were pulse-labeled for 30 min with Met35[S] after starvation in methionine-free medium for 30 min, either in the presence (25 μM) or absence of TTP-8307. Cells were lysed and translation products were analyzed by SDS-polyacrylamide gel electrophoresis. The data represent averages ± the SD from three independent experiments.
FIG. 4.
Plaque phenotypes of wild-type CVB3 and recombinant clones.
FIG. 5.
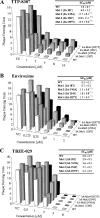
Viral replication of wild-type and recombinant viruses in the presence of TTP-8307, enviroxime and TBZE-029. Each graph represents the number of PFU produced by wild-type CVB3 or by any of the four constructed 3A mutants—3A[I8T], 3A[V45A], 3A[I54F], and 3A[H57Y]—at a given compound concentration. Calculated EC90 values represent averages ± the SD from five independent experiments. **, P < 0.05.
FIG. 6.
(A) Sequence alignment of 3A proteins from CVB3, PV1, HRV2, and HRV14. The residues that are circled were earlier identified as amino acid mutations in enviroxime (or analog)-resistant variants (3, 22, 23). The arrows indicate mutations in TTP-8307 resistant CVB3, identified in the present study. The residues between brackets form a 22-residue hydrophobic region involved in membrane anchoring of 3A/3AB. The region underlined in green is suggested to be a “hot spot” for viral mutations when virus is selected in the presence of TTP-8307 or enviroxime. (B) Predicted solution structure of the N-terminal soluble domain of the PV 3A protein. The two cylinders represent two α-helices that form an α-helical hairpin. Adapted from Wessels et al. (46) with permission. (C) Location of the three predominant amino acid mutations involved in CVB3 resistance to TTP-8307 in a structural model of protein 3A.
Similar articles
- Coxsackievirus mutants that can bypass host factor PI4KIIIβ and the need for high levels of PI4P lipids for replication.
van der Schaar HM, van der Linden L, Lanke KH, Strating JR, Pürstinger G, de Vries E, de Haan CA, Neyts J, van Kuppeveld FJ. van der Schaar HM, et al. Cell Res. 2012 Nov;22(11):1576-92. doi: 10.1038/cr.2012.129. Epub 2012 Sep 4. Cell Res. 2012. PMID: 22945356 Free PMC article. - The thiazolobenzimidazole TBZE-029 inhibits enterovirus replication by targeting a short region immediately downstream from motif C in the nonstructural protein 2C.
De Palma AM, Heggermont W, Lanke K, Coutard B, Bergmann M, Monforte AM, Canard B, De Clercq E, Chimirri A, Pürstinger G, Rohayem J, van Kuppeveld F, Neyts J. De Palma AM, et al. J Virol. 2008 May;82(10):4720-30. doi: 10.1128/JVI.01338-07. Epub 2008 Mar 12. J Virol. 2008. PMID: 18337578 Free PMC article. - Modulation of proteolytic polyprotein processing by coxsackievirus mutants resistant to inhibitors targeting phosphatidylinositol-4-kinase IIIβ or oxysterol binding protein.
Lyoo H, Dorobantu CM, van der Schaar HM, van Kuppeveld FJM. Lyoo H, et al. Antiviral Res. 2017 Nov;147:86-90. doi: 10.1016/j.antiviral.2017.10.006. Epub 2017 Oct 9. Antiviral Res. 2017. PMID: 29024767 - Antiviral agents against picornaviruses.
Eggers HJ. Eggers HJ. Antiviral Res. 1985;Suppl 1:57-65. doi: 10.1016/s0166-3542(85)80009-7. Antiviral Res. 1985. PMID: 3002267 Review. No abstract available.
Cited by
- Direct-Acting Antivirals and Host-Targeting Approaches against Enterovirus B Infections: Recent Advances.
Tammaro C, Guida M, Appetecchia F, Biava M, Consalvi S, Poce G. Tammaro C, et al. Pharmaceuticals (Basel). 2023 Jan 29;16(2):203. doi: 10.3390/ph16020203. Pharmaceuticals (Basel). 2023. PMID: 37259352 Free PMC article. Review. - Evaluation of 3D Human Intestinal Organoids as a Platform for EV-A71 Antiviral Drug Discovery.
Masmoudi F, Santos-Ferreira N, Pajkrt D, Wolthers KC, DeGroot J, Vlaming MLH, Rocha-Pereira J, Buti L. Masmoudi F, et al. Cells. 2023 Apr 12;12(8):1138. doi: 10.3390/cells12081138. Cells. 2023. PMID: 37190047 Free PMC article. - Characterization of Anti-Poliovirus Compounds Isolated from Edible Plants.
Arita M, Fuchino H. Arita M, et al. Viruses. 2023 Mar 31;15(4):903. doi: 10.3390/v15040903. Viruses. 2023. PMID: 37112883 Free PMC article. - Hand, Foot, and Mouth Disease Challenges and Its Antiviral Therapeutics.
Li Z, Ji W, Chen S, Duan G, Jin Y. Li Z, et al. Vaccines (Basel). 2023 Mar 1;11(3):571. doi: 10.3390/vaccines11030571. Vaccines (Basel). 2023. PMID: 36992155 Free PMC article. - Structure-function analysis of enterovirus protease 2A in complex with its essential host factor SETD3.
Peters CE, Schulze-Gahmen U, Eckhardt M, Jang GM, Xu J, Pulido EH, Bardine C, Craik CS, Ott M, Gozani O, Verba KA, Hüttenhain R, Carette JE, Krogan NJ. Peters CE, et al. Nat Commun. 2022 Sep 8;13(1):5282. doi: 10.1038/s41467-022-32758-3. Nat Commun. 2022. PMID: 36075902 Free PMC article.
References
- Aldabe, R., A. Barco, and L. Carrasco. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271:23134-23137. - PubMed
- Brown-Augsburger, P., L. M. Vance, S. K. Malcolm, H. Hsiung, D. P. Smith, and B. A. Heinz. 1999. Evidence that enviroxime targets multiple components of the rhinovirus 14 replication complex. Arch. Virol. 144:1569-1585. - PubMed
- Choe, S. S., D. A. Dodd, and K. Kirkegaard. 2005. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology 337:18-29. - PubMed
- Couzin, J. 2006. Report concludes polio drugs are needed—after disease is eradicated. Science 311:1539. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources