Small RNAs as guardians of the genome - PubMed (original) (raw)
Review
Small RNAs as guardians of the genome
Colin D Malone et al. Cell. 2009.
Abstract
Transposons populate the landscape of all eukaryotic genomes. Often considered purely genomic parasites, transposons can also benefit their hosts, playing roles in gene regulation and in genome organization and evolution. Peaceful coexistence with mobile elements depends upon adaptive control mechanisms, since unchecked transposon activity can impact long-term fitness and acutely reduce the fertility of progeny. Here, we review the conserved roles played by small RNAs in the adaptation of eukaryotes to coexist with their genomic colonists. An understanding of transposon-defense pathways has uncovered recurring themes in the mechanisms by which genomes distinguish "self" from "non-self" and selectively silence the latter.
Figures
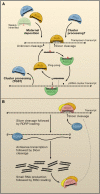
Figure 1. Strategies for Shaping Small RNA Populations
Ping-pong and RNA-dependent RNA polymerase (RDRP) cycles amplify small RNA populations against target molecules. (A) Maternal, and possibly primary, cluster-derived piRNAs initiate the ping-pong cycle by targeting the cleavage of transposon transcripts. A secondary piRNA is then produced after a 3′ cleavage of unknown source. AGO3 loads secondary piRNAs, which then target the cleavage of antisense transposon transcripts from piRNA clusters. A novel piRNA is subsequently produced and loaded into Aub, which can then target an additional transposon transcript, restarting the amplification cycle. (TGS, transcriptional gene silencing.) (B) Transcripts targeted by small RNAs are cleaved by the RNA-induced silencing complex (RISC). RDRP subsequently loads and reverse-transcribes the transcript. The new dsRNA molecule is recognized and processed into small RNAs by a Dicer protein, followed by loading of single-stranded small RNAs into RISC. Finally, RISC is able to target additional cellular transcripts and restart the cycle.
Figure 2. Model of RNAi-Based Scanning and Sequence Elimination in Tetrahymena thermophila
Simplified view of Twi1p scanning and targeting of sequence elimination. Nuclear development is not depicted (see Matzke and Birchler, 2005, for a detailed overview of nuclear progression during development). Each depicted cell is one of a mating pair (shaded cells). (A) Generation of scan RNAs (scnRNAs) from the eliminated (green) and noneliminated (orange) germline sequences. (B) Micronuclear export and loading of scnRNAs into cytoplasmic Twi1p. (C) Twi1p import into the parental macronucleus, where Twi1p with bound scnRNAs “scans” the parental genome and are depleted from the population if matching to the parental, rearranged genome. (D) Export of Twi1p, bound with eliminated-element-enriched scnRNAs, to the cytoplasm. (E) Twi1p import into the developing macronucleus followed by targeting of elements to be eliminated with histone modifications (me, methylation). (F) The newly rearranged macronuclear genome after recognition and elimination of elements targeted by the rearrangement machinery.
Similar articles
- [Regulation of fertility by the small RNA pathway that defends the genome against transposons].
Shi M, Tang AF, Cai ZM. Shi M, et al. Yi Chuan. 2010 Jan;32(1):11-6. doi: 10.3724/sp.j.1005.2010.00011. Yi Chuan. 2010. PMID: 20085880 Review. Chinese. - The eukaryotic genome as an RNA machine.
Amaral PP, Dinger ME, Mercer TR, Mattick JS. Amaral PP, et al. Science. 2008 Mar 28;319(5871):1787-9. doi: 10.1126/science.1155472. Science. 2008. PMID: 18369136 - [Advances in mechanism of small RNAs].
Xie ZH. Xie ZH. Yi Chuan. 2009 Dec;31(12):1205-13. doi: 10.3724/sp.j.1005.2009.01205. Yi Chuan. 2009. PMID: 20042387 Review. Chinese. - [The Role of Transposable Elements in Ontogenesis].
Mustafin RN, Khusnutdinova EK. Mustafin RN, et al. Usp Fiziol Nauk. 2016 Jul-Sep;47(3):70-96. Usp Fiziol Nauk. 2016. PMID: 29283231 Review. Russian. - The long and the short of noncoding RNAs.
Brosnan CA, Voinnet O. Brosnan CA, et al. Curr Opin Cell Biol. 2009 Jun;21(3):416-25. doi: 10.1016/j.ceb.2009.04.001. Epub 2009 May 15. Curr Opin Cell Biol. 2009. PMID: 19447594 Review.
Cited by
- The mechanism of ageing: primary role of transposable elements in genome disintegration.
Sturm Á, Ivics Z, Vellai T. Sturm Á, et al. Cell Mol Life Sci. 2015 May;72(10):1839-47. doi: 10.1007/s00018-015-1896-0. Epub 2015 Apr 3. Cell Mol Life Sci. 2015. PMID: 25837999 Free PMC article. - Mechanisms of Evolutionary Innovation Point to Genetic Control Logic as the Key Difference Between Prokaryotes and Eukaryotes.
Bains W, Schulze-Makuch D. Bains W, et al. J Mol Evol. 2015 Aug;81(1-2):34-53. doi: 10.1007/s00239-015-9688-6. Epub 2015 Jul 25. J Mol Evol. 2015. PMID: 26208881 - Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines.
Ishizu H, Siomi H, Siomi MC. Ishizu H, et al. Genes Dev. 2012 Nov 1;26(21):2361-73. doi: 10.1101/gad.203786.112. Genes Dev. 2012. PMID: 23124062 Free PMC article. Review. - Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9.
Lu X, Wontakal SN, Kavi H, Kim BJ, Guzzardo PM, Emelyanov AV, Xu N, Hannon GJ, Zavadil J, Fyodorov DV, Skoultchi AI. Lu X, et al. Science. 2013 Apr 5;340(6128):78-81. doi: 10.1126/science.1234654. Science. 2013. PMID: 23559249 Free PMC article. - Primordial germ cells in mice.
Saitou M, Yamaji M. Saitou M, et al. Cold Spring Harb Perspect Biol. 2012 Nov 1;4(11):a008375. doi: 10.1101/cshperspect.a008375. Cold Spring Harb Perspect Biol. 2012. PMID: 23125014 Free PMC article. Review.
References
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. - PubMed
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. - PubMed
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources