Selective stabilization of mammalian microRNAs by 3' adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2 - PubMed (original) (raw)
Selective stabilization of mammalian microRNAs by 3' adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2
Takayuki Katoh et al. Genes Dev. 2009.
Abstract
The steady-state levels of microRNAs (miRNAs) and their activities are regulated by the post-transcriptional processes. It is known that 3' ends of several miRNAs undergo post-dicing adenylation or uridylation. We isolated the liver-specific miR-122 from human hepatocytes and mouse livers. Direct analysis by mass spectrometry revealed that one variant of miR-122 has a 3'-terminal adenosine that is introduced after processing by Dicer. We identified GLD-2, which is a regulatory cytoplasmic poly(A) polymerase, as responsible for the 3'-terminal adenylation of miR-122 after unwinding of the miR-122/miR-122* duplex. In livers from GLD-2-null mice, the steady-state level of the mature form of miR-122 was specifically lower than in heterozygous mice, whereas no reduction of pre-miR-122 was observed, demonstrating that 3'-terminal adenylation by GLD-2 is required for the selective stabilization of miR-122 in the liver.
Figures
Figure 1.
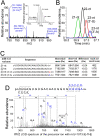
Mass spectrometric characterization of miR-122 isolated from mouse liver. (A) Mass spectrum of the 23-nt (3′-A) variant of miR-122. The multiply charged negative ions, [M-8H]8−, [M-9H]9−, and [M-10H]10−, of deprotonated miR-122 can be seen. (Inset) A series of isotopic ions of [M-10H]10− are shown in the magnified graph; the monoisotopic ion (m/z 751.19016) is indicated. The mass spectrum was acquired in the range of m/z 750–1500 with mass resolution of 30,000 (FWHM). (B) Mass chromatograms shown by [M-8H]8− and [M-9H]9− of each miR-122 variant, 21-nt (green), 22-nt (black), 23-nt (3′-U), (red) and23-nt (3′-A) (blue). (C) Sequences and exact molecular masses of the miR-122 variants. The read number from the pyrosequencing for each variant is indicated. The total number of reads was 16,638. (D) CID spectrum of the 23-nt (3′-A) variant of miR-122. [M-7H]7− (m/z 1073.9) was selected as a precursor mass for CID. The product ions were assigned according to Mcluckey et al. (1992). Partial sequences from both termini determined by this spectrum are indicated.
Figure 2.
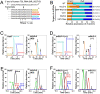
GLD-2 is a 3′ adenylase for cytoplasmic ncRNAs. (A) 3′-End sequence of human 7SL RNA gene (NR_002715) and its transcribed sequences with 3′-terminal variations. Cleavage sites of RNase T1 are shown by arrowheads. The five different 3′-terminal fragments are colored yellow (UCUCU), orange (UCUCUu), red (UCUCUuu), light blue (UCUCUa), and blue (UCUCUua). Lowercase letters (u or a) stand for additional nucleosides (uridine or adenosine) attached at the 3′ ends after transcription. 3′-Terminal fragments bearing 2′,3′-cyclic phosphates are not shown. (B) Composition and ratio of 3′ termini of 7SL RNAs isolated from Huh7 cells when each indicated nucleotidyl-transferase was knocked down. The steady-state level of each mRNA in the shRNA treated cells is shown in parentheses. The relative abundance of 3′-terminal fragments was calculated from the intensity of the mass chromatogram for each fragment. The color code of each fragment is the same as in A. (C) Mass chromatograms of doubly charged ions of five different 3′-terminal fragments of 7SL RNAs from Huh7 cells transfected (right panel) or not transfected (left panel) with an shRNA targeting to hGLD-2: UCUCU (m/z 732.00–732.75, yellow), UCUCUu (m/z 885.00–885.75, orange), UCUCUuu (m/z 1038.00–1038.75, red), UCUCUa (m/z 896.50–897.25, light blue), and UCUCUua (m/z 1049.50–1050.25, blue). (D) Mass chromatograms of doubly charged ions of four different 3′-terminal fragments of 7SL RNAs from livers of mGLD-2+/− mice (left panel) or mGLD-2−/− mice (right panel). The color code and m/z value of each fragment are the same as in A–C. (E) Mass chromatograms of the [M-8H]8−, [M-9H]9−, and [M-10H]10− ions of each miR-122 variant [21-nt (MW 6847.84, green), 22-nt (MW 7192.88, black), 23-nt (3′-U) (MW 7498.91, red), and 23-nt (3′-A) (MW 7521.94, blue)] from Huh7 cells transfected (right panel) or not transfected (left panel) with an shRNA targeting hGLD-2. (F) Mass chromatograms of the [M-8H]8−, [M-9H]9−, and [M-10H]10− ions of each miR-122 variant from livers ofmGLD-2+/− mice (left panel) or mGLD-2−/− mice (right panel). The color code and MW value of each variant are the same as in E.
Figure 3.
Steady-state level of miR-122 is markedly decreased in mGLD-2 knockout mice. (A) Northern blotting detecting miR-122 (left panel) and miR-21 (right panel) from mGLD-2+/− and mGLD-2−/− mice. As positive controls, 40 fmol each of synthetic miR-122 (22-nt and 23-nt) and miR-21 (21-nt) was run along with the total RNAs. Northern blotting of mitochondrial tRNAAla was used as a loading control for each miRNA. (B) Ratio of the steady-state level of each miRNA from mGLD-2−/− to that from mGLD-2+/−, as measured by real-time RT–PCR. Each ratio was normalized by the ratio of miR-21 (whose value was defined as 1). (C) Ratio of the steady-state level of each pre-miRNA from mGLD-2−/− to that from mGLD-2+/− as measured by real-time RT–PCR. Each ratio was normalized by the ratio of pre-miR-21 (whose value was defined as 1). (D) Expression levels of miR-122-target genes (AldoA, bckdk, cd320, Cyclin G1, and Ndrg3) and GAPDH mRNA in mGLD-2−/− relative to those in mGLD-2+/− were measured by real-time RT–PCR. The relative expression level of each mRNA was normalized by the relative ratio of ACTB mRNA (whose value was defined as 1). The data in B–D are shown as values ± SD and reflect the averages of six independent experiments.
Figure 4.
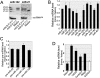
In vitro 3′ adenylation of miR-122 by immunoprecipitated mGLD-2. (A) The synthetic miR-122 was 3′-adenylated in vitro by the immunoprecipitated mGLD-2 in the presence of [α-32P]ATP. Wild-type mGLD-2 (mGLD-2 WT) or its inactive mutant (mGLD-2 D215A) was immunoprecipitated with anti-Flag M2-agarose beads from the lysate of Huh7 cells transfected with pcDNA3 Flag-HA-mGLD-2 or its inactivated mutant form (pcDNA3 Flag-HA-mGLD-2 D215A). No vector represents a negative control preparation from the same cell lysate untransfected. [α-32P]AMP-labeled miR-122 was analyzed by 20% denaturing PAGE and visualized by an imaging analyzer (BAS5000; FujiFilm). The marker is the 5′-32P-labeled miR-122 (22-nt). (B) The synthetic miR-122 (single strand) and miR-122/mi-R122* duplex were 3′-adenylated in vitro by the immunoprecipitated mGLD-2 in the presence of [α-32P]ATP. The products were analyzed as described in A.
Figure 5.
Proposed model for the selective stabilization of miR-122 by 3′ nucleotide addition and exonucleolytic degradation. In the cytoplasm, pre-miR-122 is processed by Dicer into mature miR-122 of a uniform 22-nt length (red letters). After unwinding of the miR-122/miR-122* duplex, the 3′ terminus of miR-122 is elongated by the addition of uridine/adenosine by a putative 3′ uridylase or GLD-2 and shortened to 21 nt or less by a putative 3′–5′ exonuclease. Competition between the addition and removal of 3′ nucleotides might determine the steady-state level of miR-122. The average size of miR-122 should be determined by the fact that 21 to 23 nt of miRNAs are preferentially loaded into Argonaute 2 in the RISC machinery.
Similar articles
- The RNA-binding protein QKI-7 recruits the poly(A) polymerase GLD-2 for 3' adenylation and selective stabilization of microRNA-122.
Hojo H, Yashiro Y, Noda Y, Ogami K, Yamagishi R, Okada S, Hoshino SI, Suzuki T. Hojo H, et al. J Biol Chem. 2020 Jan 10;295(2):390-402. doi: 10.1074/jbc.RA119.011617. Epub 2019 Dec 1. J Biol Chem. 2020. PMID: 31792053 Free PMC article. - Adenylation of maternally inherited microRNAs by Wispy.
Lee M, Choi Y, Kim K, Jin H, Lim J, Nguyen TA, Yang J, Jeong M, Giraldez AJ, Yang H, Patel DJ, Kim VN. Lee M, et al. Mol Cell. 2014 Dec 4;56(5):696-707. doi: 10.1016/j.molcel.2014.10.011. Epub 2014 Nov 13. Mol Cell. 2014. PMID: 25454948 Free PMC article. - Destabilization of microRNAs in human cells by 3' deadenylation mediated by PARN and CUGBP1.
Katoh T, Hojo H, Suzuki T. Katoh T, et al. Nucleic Acids Res. 2015 Sep 3;43(15):7521-34. doi: 10.1093/nar/gkv669. Epub 2015 Jun 30. Nucleic Acids Res. 2015. PMID: 26130707 Free PMC article. - Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)-polymerase GLD-2 by GLD-3.
Nakel K, Bonneau F, Eckmann CR, Conti E. Nakel K, et al. Proc Natl Acad Sci U S A. 2015 Jul 14;112(28):8614-9. doi: 10.1073/pnas.1504648112. Epub 2015 Jun 29. Proc Natl Acad Sci U S A. 2015. PMID: 26124149 Free PMC article. - Non-canonical poly(A) polymerase in mammalian gametogenesis.
Kashiwabara S, Nakanishi T, Kimura M, Baba T. Kashiwabara S, et al. Biochim Biophys Acta. 2008 Apr;1779(4):230-8. doi: 10.1016/j.bbagrm.2008.01.004. Epub 2008 Feb 12. Biochim Biophys Acta. 2008. PMID: 18294465 Review.
Cited by
- HER2-encoded mir-4728 forms a receptor-independent circuit with miR-21-5p through the non-canonical poly(A) polymerase PAPD5.
Newie I, Søkilde R, Persson H, Jacomasso T, Gorbatenko A, Borg Å, de Hoon M, Pedersen SF, Rovira C. Newie I, et al. Sci Rep. 2016 Oct 18;6:35664. doi: 10.1038/srep35664. Sci Rep. 2016. PMID: 27752128 Free PMC article. - Selective Suppression of the Splicing-Mediated MicroRNA Pathway by the Terminal Uridyltransferase Tailor.
Bortolamiol-Becet D, Hu F, Jee D, Wen J, Okamura K, Lin CJ, Ameres SL, Lai EC. Bortolamiol-Becet D, et al. Mol Cell. 2015 Jul 16;59(2):217-28. doi: 10.1016/j.molcel.2015.05.034. Epub 2015 Jul 2. Mol Cell. 2015. PMID: 26145174 Free PMC article. - Regulation of small RNA stability: methylation and beyond.
Ji L, Chen X. Ji L, et al. Cell Res. 2012 Apr;22(4):624-36. doi: 10.1038/cr.2012.36. Epub 2012 Mar 13. Cell Res. 2012. PMID: 22410795 Free PMC article. Review. - Secretion of microvesicular miRNAs in cellular and organismal aging.
Weilner S, Schraml E, Redl H, Grillari-Voglauer R, Grillari J. Weilner S, et al. Exp Gerontol. 2013 Jul;48(7):626-33. doi: 10.1016/j.exger.2012.11.017. Epub 2012 Dec 30. Exp Gerontol. 2013. PMID: 23283304 Free PMC article. Review. - Soma-germline interactions that influence germline proliferation in Caenorhabditis elegans.
Korta DZ, Hubbard EJ. Korta DZ, et al. Dev Dyn. 2010 May;239(5):1449-59. doi: 10.1002/dvdy.22268. Dev Dyn. 2010. PMID: 20225254 Free PMC article. Review.
References
- Barnard D.C., Ryan K., Manley J.L., Richter J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. - PubMed
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. - PubMed
- Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M.A., Xu C., Mason W.S., Moloshok T., Bort R., et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases