Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice - PubMed (original) (raw)
Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice
Ryan L Boudreau et al. Mol Ther. 2009 Jun.
Abstract
Huntington's disease (HD) is a fatal neurodegenerative disease caused by mutant huntingtin (htt) protein, and there are currently no effective treatments. Recently, we and others demonstrated that silencing mutant htt via RNA interference (RNAi) provides therapeutic benefit in HD mice. We have since found that silencing wild-type htt in adult mouse striatum is tolerated for at least 4 months. However, given the role of htt in various cellular processes, it remains unknown whether nonallele-specific silencing of both wild-type and mutant htt is a viable therapeutic strategy for HD. Here, we tested whether cosilencing wild-type and mutant htt provides therapeutic benefit and is tolerable in HD mice. After treatment, HD mice showed significant reductions in wild-type and mutant htt, and demonstrated improved motor coordination and survival. We performed transcriptional profiling to evaluate the effects of reducing wild-type htt in adult mouse striatum. We identified gene expression changes that are concordant with previously described roles for htt in various cellular processes. Also, several abnormally expressed transcripts associated with early-stage HD were differentially expressed in our studies, but intriguingly, those involved in neuronal function changed in opposing directions. Together, these encouraging and surprising findings support further testing of nonallele-specific RNAi therapeutics for HD.
Figures
**Figure 1
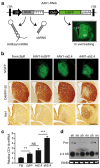
Mi2.4 demonstrates improved safety, relative to sh2.4, in HD-N171-82Q mice. (a) Diagram of recombinant AAV2/1 viral vectors which express htt-specific RNAi and hrGFP; the latter allows tracking of in vivo transduction within mouse brain (Str = striatum; LV = lateral ventricle). (b) HD-N171-82Q mice were injected with either formulation buffer (FB) or viral vectors into the striatum, and 3 months later, histological analyses were performed to assess for striatal toxicity. Photomicrographs representing hrGFP autofluorescence and immunohistochemical staining of DARPP-32-positive neurons and IbaI-positive microglia are shown for each treatment group. Scale bar = 500 µm for each photomicrograph. (c) Microglial activation was also evaluated by quantitative real-time PCR analyses measuring the endogenous mouse CD11b mRNA levels in striatal RNA samples. Normalized results are shown as mean ± SEM (n ≥ 5; ***, ** and NS indicate P < 0.001, P < 0.01 and no significance, respectively). (d) Small transcript northern blot analysis was performed to assess the levels of htt-specific antisense (2.4 AS) RNA present in AAV1-RNAi-treated striata (sh = sh2.4, mi = mi2.4; n = 3 treated striata). Sh2.4 treatment yielded a build-up of unprocessed precursor RNAs (Pre-, arrowheads) in 2 of 3 samples. Ethidium bromide (EthBr) staining was performed as a loading control. hrGFP, humanized renilla green fluorescent protein; ITR, inverted terminal repeat; RNAi, RNA interference.
**Figure 2
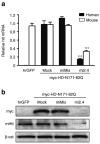
Mi2.4 silences both mutant human and wild-type mouse htt in vitro. RNAi expression plasmids were co-transfected into mouse C2C12 cells along with a plasmid expressing a myc-tagged mutant human HD-N171-82Q transgene, and gene silencing was assessed 24 hours later. (a) QPCR analyses were performed to measure the levels of mutant human and endogenous wild-type mouse htt mRNAs. Results are shown as mean ± SEM (n = 4; *** indicates P < 0.001). (b) Silencing of the human and mouse htt proteins (myc and mHtt, respectively) was assessed by western blot analyses. β-catenin (β-cat) served as the loading control. The mismatch control (miMis) contains multiple base-pair changes that render the miRNA ineffective against htt. Mock-treated cells received promoter-only plasmid (i.e., no miRNA). hrGFP-treated cells served as a control for evaluating the specificity of the human and mouse htt QPCR primer/probes and antibodies. hrGFP, humanized renilla green fluorescent protein; QPCR, quantitative real-time PCR.
**Figure 3
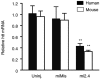
Mi2.4 silences both mutant human and wild-type mouse htt mRNAs in HD-N171-82Q mice. HD-N171-82Q mice were injected unilaterally into the striatum with AAV1-miMis, or AAV1-mi2.4, and quantitative real-time analyses were performed on striatal RNA samples to measure the levels of mutant human and wild-type mouse htt mRNAs at 4 weeks after treatment. Results are shown as mean ± SEM (n ≥ 3; ** indicates P < 0.01) relative to the uninjected contralateral striata.
**Figure 4
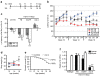
Mi2.4 provides therapeutic benefit in HD-N171-82Q mice. (a) Timeline for testing the therapeutic efficacy of AAV1-mi2.4 in HD-N171-82Q mice. HD-N171-82Q mice were injected bilaterally into the striatum with formulation buffer (FB), AAV1-hrGFP, or AAV1-mi2.4. Similarly, wild-type littermates were treated with formulation buffer. All mice were injected at 7 weeks of age, weighed and tested on the rotarod (R) apparatus at 10, 14, and 18 weeks and ultimately sacrificed (Sac) at 20 weeks for QPCR and histological analyses. (b) Rotarod data from four consecutive days at 10, 14, and 18 weeks of age is shown as latency to fall (mean ± SEM of trials 1–3 for each group per day). (c) To evaluate the progress of each group over the time-course of the therapeutic trial, the rotarod data is shown as the mean change ± SEM in average weekly performance relative to week 10 (*** and ** indicate P = 0.002 and P = 0.02, respectively). (d) The average body weights for mice in each group are shown for the indicated ages (mean ± SEM; ** represents P < 0.01). Note: legend shown in b applies. (e) Kaplan–Meier survival analysis up to 20 weeks of age (i.e., the time of killing) is shown. In parentheses is the fraction of surviving mice at the end-point per the initial total for each group. The control group consists of HD-N171-82Q mice treated with formulation buffer or AAV1-hrGFP. (f) Mice were killed at week 20, and QPCR analyses were performed on striatal RNA samples to measure mutant human and wild-type mouse htt mRNA levels. Normalized results are shown as mean ± SEM (n ≥ 5; *** and ** indicate P < 0.001 and P < 0.01, respectively). HD, Huntington's disease; hrGFP, humanized renilla green fluorescent protein; QPCR, quantitative real-time PCR; WT, wild type.
**Figure 5
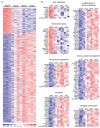
Transcriptional changes resulting from knockdown of wild-type htt in mouse striatum. (a) Heat map depicting the relative fold change in expression levels of the 473 transcripts that were differentially expressed by ≥2.0-fold (P < 0.01, 107 downregulated and 366 upregulated) between the control hrGFP-treated group and the htt-knockdown group (consisting of samples treated with mi2.4, sh2.4, or sh8.2). Data for each experimental sample (n = 3 per indicated treatment) are shown. The scale bar indicates fold change with blue and red tones signifying downregulated and upregulated, respectively. (b) Functional gene annotation clustering performed on the 107 downregulated and 366 upregulated transcripts identified enrichments in the indicated cellular components and processes. Heat maps depicting the fold change in expression of the genes within each enriched category are shown. The scale bar in a applies. hrGFP, humanized renilla green fluorescent protein.
**Figure 6
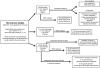
Microarray analyses workflow and summary of results. hrGFP, humanized renilla green fluorescent protein; RNAi, RNA interference.
Similar articles
- Sustained effects of nonallele-specific Huntingtin silencing.
Drouet V, Perrin V, Hassig R, Dufour N, Auregan G, Alves S, Bonvento G, Brouillet E, Luthi-Carter R, Hantraye P, Déglon N. Drouet V, et al. Ann Neurol. 2009 Mar;65(3):276-85. doi: 10.1002/ana.21569. Ann Neurol. 2009. PMID: 19334076 - Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington's disease.
McBride JL, Pitzer MR, Boudreau RL, Dufour B, Hobbs T, Ojeda SR, Davidson BL. McBride JL, et al. Mol Ther. 2011 Dec;19(12):2152-62. doi: 10.1038/mt.2011.219. Epub 2011 Oct 25. Mol Ther. 2011. PMID: 22031240 Free PMC article. - RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model.
Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. Harper SQ, et al. Proc Natl Acad Sci U S A. 2005 Apr 19;102(16):5820-5. doi: 10.1073/pnas.0501507102. Epub 2005 Apr 5. Proc Natl Acad Sci U S A. 2005. PMID: 15811941 Free PMC article. - Transcriptional dysregulation of coding and non-coding genes in cellular models of Huntington's disease.
Bithell A, Johnson R, Buckley NJ. Bithell A, et al. Biochem Soc Trans. 2009 Dec;37(Pt 6):1270-5. doi: 10.1042/BST0371270. Biochem Soc Trans. 2009. PMID: 19909260 Review. - Delivering a disease-modifying treatment for Huntington's disease.
Godinho BM, Malhotra M, O'Driscoll CM, Cryan JF. Godinho BM, et al. Drug Discov Today. 2015 Jan;20(1):50-64. doi: 10.1016/j.drudis.2014.09.011. Epub 2014 Sep 26. Drug Discov Today. 2015. PMID: 25256777 Review.
Cited by
- A striatal-enriched intronic GPCR modulates huntingtin levels and toxicity.
Yao Y, Cui X, Al-Ramahi I, Sun X, Li B, Hou J, Difiglia M, Palacino J, Wu ZY, Ma L, Botas J, Lu B. Yao Y, et al. Elife. 2015 Mar 4;4:e05449. doi: 10.7554/eLife.05449. Elife. 2015. PMID: 25738228 Free PMC article. - The role of amyloidogenic protein oligomerization in neurodegenerative disease.
Lotz GP, Legleiter J. Lotz GP, et al. J Mol Med (Berl). 2013 Jun;91(6):653-64. doi: 10.1007/s00109-013-1025-1. Epub 2013 Mar 27. J Mol Med (Berl). 2013. PMID: 23529761 Review. - A brief history of triplet repeat diseases.
Budworth H, McMurray CT. Budworth H, et al. Methods Mol Biol. 2013;1010:3-17. doi: 10.1007/978-1-62703-411-1_1. Methods Mol Biol. 2013. PMID: 23754215 Free PMC article. Review. - Taking a break from huntingtin.
Davidson BL. Davidson BL. Mol Ther Nucleic Acids. 2012 Aug 21;1(8). doi: 10.1038/mtna.2012.33. Mol Ther Nucleic Acids. 2012. PMID: 23344175 Free PMC article. No abstract available. - Taking a break from huntingtin.
Davidson BL. Davidson BL. Mol Ther. 2012 Oct;20(10):1838. doi: 10.1038/mt.2012.191. Mol Ther. 2012. PMID: 23023055 Free PMC article. No abstract available.
References
- Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004;15:95–105. - PubMed
- Saudou F, Finkbeiner S, Devys D., and , Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. - PubMed
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. - PubMed
- Zhai W, Jeong H, Cui L, Krainc D., and , Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. - PubMed
- Qiu Z, Norflus F, Singh B, Swindell MK, Buzescu R, Bejarano M, et al. Sp1 is up-regulated in cellular and transgenic models of Huntington disease, and its reduction is neuroprotective. J Biol Chem. 2006;281:16672–16680. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HD-44093/HD/NICHD NIH HHS/United States
- T32 HL007121/HL/NHLBI NIH HHS/United States
- P01 NS050210/NS/NINDS NIH HHS/United States
- DK-54759/DK/NIDDK NIH HHS/United States
- NS-50210/NS/NINDS NIH HHS/United States
- R01 HD044093/HD/NICHD NIH HHS/United States
- P30 DK054759/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical