Structural basis of the interaction between integrin alpha6beta4 and plectin at the hemidesmosomes - PubMed (original) (raw)
Structural basis of the interaction between integrin alpha6beta4 and plectin at the hemidesmosomes
José M de Pereda et al. EMBO J. 2009.
Abstract
The interaction between the integrin alpha6beta4 and plectin is essential for the assembly and stability of hemidesmosomes, which are junctional adhesion complexes that anchor epithelial cells to the basement membrane. We describe the crystal structure at 2.75 A resolution of the primary alpha6beta4-plectin complex, formed by the first pair of fibronectin type III domains and the N-terminal region of the connecting segment of beta4 and the actin-binding domain of plectin. Two missense mutations in beta4 (R1225H and R1281W) linked to nonlethal forms of epidermolysis bullosa prevent essential intermolecular contacts. We also present two structures at 1.75 and 2.05 A resolution of the beta4 moiety in the absence of plectin, which reveal a major rearrangement of the connecting segment of beta4 on binding to plectin. This conformational switch is correlated with the way alpha6beta4 promotes stable adhesion or cell migration and suggests an allosteric control of the integrin.
Figures
Figure 1
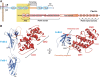
Structure of the integrin β4–plectin complex. (A) Schematic representation of the domain organisation of plectin and the cytoplasmic domains of integrin α6β4. The regions of both proteins corresponding to the primary interaction site, whose crystal structure is presented herein, are highlight by a yellow box. (B) Ribbon representation in two orthogonal views of the integrin β4 (residues 1127–1318 in blue and 1324–1330 in orange) bound to plectin (residues 60–290 in red), with secondary structure elements labelled. Molecular figures were prepared using PyMOL (Delano, 2002).
Figure 2
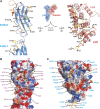
Integrin β4–plectin interface. (A) Open-book view of the complex. Ribbon representation of the β4 (left) and plectin (right) structure with residues that constitute the interface shown as sticks. (B, C) Surface representation coloured by electrostatic potential (from -12 kT/e in red to 12 kT/e in blue) of β4 (B) and plectin (C) in the same orientation as in (A). Residues that form part of the interfaces are labelled on each surface. The backbone of the companion partner is partially shown as a wire, with the side chains that participate in the interaction shown as sticks and labelled with arrows.
Figure 3
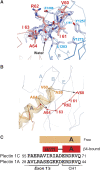
Structure of the N-terminal arm of plectin. (A) Representation of the N-terminal arm of plectin (red) bound to the FnIII-2 of β4 (blue). An omit map (2mFobs-DFcalc, contoured at 1σ) is shown in blue; the map was calculated after simulated annealing refinement of a model in which the plectin arm was omitted. (B) Cα-trace of the N-terminal region of ABD in the absence of plectin (orange, PDB 1MB8) superimposed on the structure of the ABD (red) bound to β4 (blue). (C) Comparison of the sequences of plectin 1C and 1A upstream of the ABD. Only the initial residues of the CH1, which is common to the two isoforms, are shown. The secondary structure elements in the free (orange) and β4-bound (red) structures of the ABD are shown on top.
Figure 4
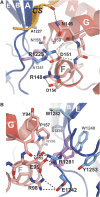
Structural basis of missense mutations of β4 linked to epidermolysis bullosa. (A) Close-up showing the intermolecular contacts between R1225 of β4 (blue) and plectin (red). A hypothetical conformation of the His side chain in the R1225H mutant is shown to illustrate the effect of this substitution, which prevents the interactions with N146, R148 and D151 in plectin. (B) Detailed view of the contacts established by R1281 of β4, and nearby residues (coloured as in A). A possible structure of the Trp side chain in the disease-linked mutation R1281W is shown. The imidazole ring does not maintain the H-bonds established by R1281 and most likely it causes steric clashes with W1240 of β4 and P157 of plectin.
Figure 5
Conformational change of the connecting segment of β4 between the free and plectin-bound structures. (A) Ribbon representation of the 1126–1348 free β4 structure, with the FnIII-2 in blue and the CS in orange (the FnIII-1 has been omitted in all panels for clarity). The Pro-rich loop (residues 1324–1338), for which no electron density was observed, is shown as a dotted line. (B) Representation of the 1126–1339 free β4 structure in the same orientation and colouring as in (A). (C) Detailed view of the CS (shown as sticks, orange) in the 1126–1339 free β4 structure. P1323, P1327 and P1330 are highly exposed to the solvent, whereas P1333 contact the FnIII-2 right before the β-strand H. (D) Structure of the CS in the complex of β4 (blue) with plectin (red). The view is in the same orientation as in panels A and B, and the CH2 was omitted for clarity. (E) Schematic representation of the FnIII-2 and the CS of β4 in the free (left) and plectin-bound (right) structures. The arrows illustrate the structural reorganisation of the CS induced by plectin binding.
Similar articles
- Two different mutations in the cytoplasmic domain of the integrin beta 4 subunit in nonlethal forms of epidermolysis bullosa prevent interaction of beta 4 with plectin.
Koster J, Kuikman I, Kreft M, Sonnenberg A. Koster J, et al. J Invest Dermatol. 2001 Dec;117(6):1405-11. doi: 10.1046/j.0022-202x.2001.01567.x. J Invest Dermatol. 2001. PMID: 11886501 - Advances and perspectives of the architecture of hemidesmosomes: lessons from structural biology.
de Pereda JM, Ortega E, Alonso-García N, Gómez-Hernández M, Sonnenberg A. de Pereda JM, et al. Cell Adh Migr. 2009 Oct-Dec;3(4):361-4. doi: 10.4161/cam.3.4.9525. Epub 2009 Oct 16. Cell Adh Migr. 2009. PMID: 19736524 Free PMC article. Review. - Plectin isoform-dependent regulation of keratin-integrin alpha6beta4 anchorage via Ca2+/calmodulin.
Kostan J, Gregor M, Walko G, Wiche G. Kostan J, et al. J Biol Chem. 2009 Jul 3;284(27):18525-36. doi: 10.1074/jbc.M109.008474. Epub 2009 May 6. J Biol Chem. 2009. PMID: 19419971 Free PMC article. - Dynamics of the alpha6beta4 integrin in keratinocytes.
Geuijen CA, Sonnenberg A. Geuijen CA, et al. Mol Biol Cell. 2002 Nov;13(11):3845-58. doi: 10.1091/mbc.02-01-0601. Mol Biol Cell. 2002. PMID: 12429829 Free PMC article. - Regulation of hemidesmosome disassembly by growth factor receptors.
Margadant C, Frijns E, Wilhelmsen K, Sonnenberg A. Margadant C, et al. Curr Opin Cell Biol. 2008 Oct;20(5):589-96. doi: 10.1016/j.ceb.2008.05.001. Epub 2008 Jun 24. Curr Opin Cell Biol. 2008. PMID: 18583123 Review.
Cited by
- Plectin: Dual Participation in Tumor Progression.
Wang Z, Wang W, Luo Q, Song G. Wang Z, et al. Biomolecules. 2024 Aug 24;14(9):1050. doi: 10.3390/biom14091050. Biomolecules. 2024. PMID: 39334817 Free PMC article. Review. - Integrins as the pivotal regulators of cisplatin response in tumor cells.
Nasimi Shad A, Moghbeli M. Nasimi Shad A, et al. Cell Commun Signal. 2024 May 13;22(1):265. doi: 10.1186/s12964-024-01648-0. Cell Commun Signal. 2024. PMID: 38741195 Free PMC article. Review. - Prognostic value of the immunohistochemical score based on four markers in head and neck squamous cell carcinoma.
Xu QQ, Li QJ, Xu Z, Lan LL, Hou Z, Liu J, Lu L, Chen YY, Chen RZ, Wen X. Xu QQ, et al. Front Immunol. 2023 Feb 22;14:1076890. doi: 10.3389/fimmu.2023.1076890. eCollection 2023. Front Immunol. 2023. PMID: 36911694 Free PMC article. - Cytoskeletal and Cytoskeleton-Associated Proteins: Key Regulators of Cancer Stem Cell Properties.
Li Y, Wang D, Ge H, Güngör C, Gong X, Chen Y. Li Y, et al. Pharmaceuticals (Basel). 2022 Nov 8;15(11):1369. doi: 10.3390/ph15111369. Pharmaceuticals (Basel). 2022. PMID: 36355541 Free PMC article. Review. - Autosomal recessive inheritance of a novel missense mutation of ITGB4 for Epidermolysis-Bullosa pyloric-atresia: a case report.
Paine SK, Das S, Bhattacharyya C, Biswas NK, Rao R, De A, Basu A. Paine SK, et al. Mol Genet Genomics. 2022 Nov;297(6):1581-1586. doi: 10.1007/s00438-022-01941-y. Epub 2022 Aug 23. Mol Genet Genomics. 2022. PMID: 35997841
References
- Afonine PV, Grosse-Kunstleve RW, Adams PD (2005) The Phenix refinement framework. CCP4 Newsl 42, contribution 8.
- Andra K, Kornacker I, Jorgl A, Zorer M, Spazierer D, Fuchs P, Fischer I, Wiche G (2003) Plectin-isoform-specific rescue of hemidesmosomal defects in plectin (−/−) keratinocytes. J Invest Dermatol 120: 189–197 - PubMed
- Delano WL (2002) The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases