An improved ontological representation of dendritic cells as a paradigm for all cell types - PubMed (original) (raw)
An improved ontological representation of dendritic cells as a paradigm for all cell types
Anna Maria Masci et al. BMC Bioinformatics. 2009.
Abstract
Background: Recent increases in the volume and diversity of life science data and information and an increasing emphasis on data sharing and interoperability have resulted in the creation of a large number of biological ontologies, including the Cell Ontology (CL), designed to provide a standardized representation of cell types for data annotation. Ontologies have been shown to have significant benefits for computational analyses of large data sets and for automated reasoning applications, leading to organized attempts to improve the structure and formal rigor of ontologies to better support computation. Currently, the CL employs multiple is_a relations, defining cell types in terms of histological, functional, and lineage properties, and the majority of definitions are written with sufficient generality to hold across multiple species. This approach limits the CL's utility for computation and for cross-species data integration.
Results: To enhance the CL's utility for computational analyses, we developed a method for the ontological representation of cells and applied this method to develop a dendritic cell ontology (DC-CL). DC-CL subtypes are delineated on the basis of surface protein expression, systematically including both species-general and species-specific types and optimizing DC-CL for the analysis of flow cytometry data. We avoid multiple uses of is_a by linking DC-CL terms to terms in other ontologies via additional, formally defined relations such as has_function.
Conclusion: This approach brings benefits in the form of increased accuracy, support for reasoning, and interoperability with other ontology resources. Accordingly, we propose our method as a general strategy for the ontological representation of cells. DC-CL is available from http://www.obofoundry.org.
Figures
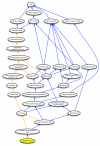
Figure 1
The representation of Langerhans cells in the Cell Ontology. A portion of the Cell Ontology is shown with ovals corresponding to cell types defined in the ontology and arrows corresponding to relations between those cell types. Langerhans cell is represented by a yellow oval; blue arrows correspond to is_a relations, and orange arrows correspond to develops_from relations. Only a subset of Langerhans cell parent types are included in the figure.
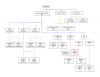
Figure 2
The representation of dendritic cell types in the Dendritic Cell Ontology (DC-CL). Rectangles correspond to the terms for dendritic cell types represented in DC-CL, and the lines connecting the rectangles correspond to the is_a relations between these cell types. Black lines connect the highest-level terms in DC-CL to the Cell Ontology term leukocyte. Blue lines connect the DC-CL term conventional dendritic cell to the terms for its subtypes, while red lines connect these terms to the respective subtype terms. The green lines connect the terms _CD11c_- plasmacytoid dendritic cell and CD11c low plasmacytoid dendritic cell to the terms for their respective subtypes. Abbreviations used in the figure are: DC, dendritic cell; PDC, plasmacytoid dendritic cell; and LC, Langerhans Cell.
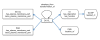
Figure 3
The ontologies and relations referred to in the Dendritic Cell Ontology (DC-CL). The rectangles and ovals represent ontologies, and the arrows represent relations joining terms in the ontologies. Abbreviations for the ontology names are shown in normal font, and the relations used to link DC-CL to each ontology are shown in italics. The black arrow indicates relations used to join DC-CL terms to other DC-CL terms, while the blue arrows indicate trans-ontological relations. Ontologies and relations shown in rectangles are used to define DC-CL types, while the ontologies and relations shown in ovals are used to make non-classificatory assertions about DC-CL types. Abbreviations used in the figure are: GO CC, Gene Ontology Cellular Component Ontology; PRO, Protein Ontology; IO, Immunology Ontology; FMA, Foundational Model of Anatomy; and GO BP, Gene Ontology Biological Process Ontology.
Similar articles
- The ontology of biological sequences.
Hoehndorf R, Kelso J, Herre H. Hoehndorf R, et al. BMC Bioinformatics. 2009 Nov 18;10:377. doi: 10.1186/1471-2105-10-377. BMC Bioinformatics. 2009. PMID: 19919720 Free PMC article. - Logical development of the cell ontology.
Meehan TF, Masci AM, Abdulla A, Cowell LG, Blake JA, Mungall CJ, Diehl AD. Meehan TF, et al. BMC Bioinformatics. 2011 Jan 5;12:6. doi: 10.1186/1471-2105-12-6. BMC Bioinformatics. 2011. PMID: 21208450 Free PMC article. - Relations in biomedical ontologies.
Smith B, Ceusters W, Klagges B, Köhler J, Kumar A, Lomax J, Mungall C, Neuhaus F, Rector AL, Rosse C. Smith B, et al. Genome Biol. 2005;6(5):R46. doi: 10.1186/gb-2005-6-5-r46. Epub 2005 Apr 28. Genome Biol. 2005. PMID: 15892874 Free PMC article. - Exploitation of ontological resources for scientific literature analysis: searching genes and related diseases.
Jimeno-Yepes A, Berlanga-Llavori R, Rebholz-Schuhmann D. Jimeno-Yepes A, et al. Annu Int Conf IEEE Eng Med Biol Soc. 2009;2009:7073-8. doi: 10.1109/IEMBS.2009.5333359. Annu Int Conf IEEE Eng Med Biol Soc. 2009. PMID: 19964204 Review. - Biomedical ontologies: a functional perspective.
Rubin DL, Shah NH, Noy NF. Rubin DL, et al. Brief Bioinform. 2008 Jan;9(1):75-90. doi: 10.1093/bib/bbm059. Epub 2007 Dec 12. Brief Bioinform. 2008. PMID: 18077472 Review.
Cited by
- Protein Ontology: a controlled structured network of protein entities.
Natale DA, Arighi CN, Blake JA, Bult CJ, Christie KR, Cowart J, D'Eustachio P, Diehl AD, Drabkin HJ, Helfer O, Huang H, Masci AM, Ren J, Roberts NV, Ross K, Ruttenberg A, Shamovsky V, Smith B, Yerramalla MS, Zhang J, AlJanahi A, Çelen I, Gan C, Lv M, Schuster-Lezell E, Wu CH. Natale DA, et al. Nucleic Acids Res. 2014 Jan;42(Database issue):D415-21. doi: 10.1093/nar/gkt1173. Epub 2013 Nov 21. Nucleic Acids Res. 2014. PMID: 24270789 Free PMC article. - Top-level categories of constitutively organized material entities--suggestions for a formal top-level ontology.
Vogt L, Grobe P, Quast B, Bartolomaeus T. Vogt L, et al. PLoS One. 2011 Apr 21;6(4):e18794. doi: 10.1371/journal.pone.0018794. PLoS One. 2011. PMID: 21533043 Free PMC article. - Cell type discovery using single-cell transcriptomics: implications for ontological representation.
Aevermann BD, Novotny M, Bakken T, Miller JA, Diehl AD, Osumi-Sutherland D, Lasken RS, Lein ES, Scheuermann RH. Aevermann BD, et al. Hum Mol Genet. 2018 May 1;27(R1):R40-R47. doi: 10.1093/hmg/ddy100. Hum Mol Genet. 2018. PMID: 29590361 Free PMC article. - Levels and building blocks-toward a domain granularity framework for the life sciences.
Vogt L. Vogt L. J Biomed Semantics. 2019 Jan 28;10(1):4. doi: 10.1186/s13326-019-0196-2. J Biomed Semantics. 2019. PMID: 30691505 Free PMC article. - OPPL-Galaxy, a Galaxy tool for enhancing ontology exploitation as part of bioinformatics workflows.
Aranguren ME, Fernández-Breis JT, Mungall C, Antezana E, González AR, Wilkinson MD. Aranguren ME, et al. J Biomed Semantics. 2013 Jan 4;4(1):2. doi: 10.1186/2041-1480-4-2. J Biomed Semantics. 2013. PMID: 23286517 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- R01 AI077706/AI/NIAID NIH HHS/United States
- 1 R01 GM080646-01/GM/NIGMS NIH HHS/United States
- U54 HG004028/HG/NHGRI NIH HHS/United States
- 1 U 54 HG004028/HG/NHGRI NIH HHS/United States
- R01 AI068804/AI/NIAID NIH HHS/United States
- HG004028-01/HG/NHGRI NIH HHS/United States
- P41 HG002273/HG/NHGRI NIH HHS/United States
- R01 GM080646/GM/NIGMS NIH HHS/United States
- N01 AI40076)./AI/NIAID NIH HHS/United States
- U41 HG002273/HG/NHGRI NIH HHS/United States
- AI50019/AI/NIAID NIH HHS/United States
- N01 AI040076/AI/NIAID NIH HHS/United States
- N01AI50019/AI/NIAID NIH HHS/United States
- HG002273/HG/NHGRI NIH HHS/United States
- N01AI40076/AI/NIAID NIH HHS/United States
- R01 HG002273/HG/NHGRI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources