Inducible deposition of the histone variant H3.3 in interferon-stimulated genes - PubMed (original) (raw)
Inducible deposition of the histone variant H3.3 in interferon-stimulated genes
Tomohiko Tamura et al. J Biol Chem. 2009.
Abstract
The H3.3 histone variant is synthesized throughout cell cycle and deposited onto chromatin in a replication-independent manner. It is enriched in transcriptionally active regions of chromatin and is implicated in epigenetic memory. The dynamics of H3.3 deposition during transcriptional activation, however, have not been fully studied so far. Here we examined H3.3 incorporation into interferon (IFN)-stimulated genes in confluent mouse NIH3T3 cells expressing H3.3 fused to the yellow fluorescent protein (YFP). Following IFN stimulation, H3.3-YFP was rapidly incorporated into all four IFN-activated genes tested, with the highest enrichment seen in the distal end of the coding region. Surprisingly, H3.3 enrichment in the coding region continued for an extended period of time, long after transcription ceased. The promoter region, although constitutively enriched with H3.3-YFP, did not show an increase in its deposition in response to IFN stimulation. Further, although H3.3-YFP deposition stably remained in non-dividing cells for days after IFN stimulation, it was rapidly diminished in dividing cells. Lastly, we examined the role of H3.3 in IFN-stimulated transcription by a short hairpin RNA approach and found that IFN-stimulated transcription was significantly impaired in H3.3 knockdown cells. Results indicate that H3.3 plays a role in IFN-mediated transcription, and its deposition leaves a prolonged post-transcriptional mark in these genes.
Figures
FIGURE 1.
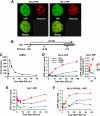
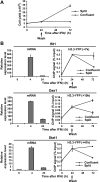
H3.3-YFP deposition at the Ifit1 gene after IFN stimulation. A, distribution of H3.3-YFP and H3.1-YFP. NIH3T3 cells were transduced with H3-YFP vectors, and YFP signals were viewed by a confocal microscopy (×600). See relative levels and chromatin association of H3-YFP in supplemental Figs. S1 and S2. B, ChIP analysis of the Ifit1 locus in IFNβ-treated NIH3T3 cells. The diagram at the top depicts the exon-intron organization. The_arrow_ indicates the transcribed region, and the gray boxes indicate exons. Positions of the primers used for ChIP were shown underneath. Cells were treated with IFNβ (100 units/ml) for 1, 3, 6, 12, 24, and 48 h. C, Ifit1 mRNA levels were measured by qRT-PCR. Transcript levels in IFN stimulation are expressed relative to those in unstimulated cells.D and E, ChIP analysis was performed with an anti-GFP antibody in NIH3T3 cells expressing H3.3-YFP- or H3.1-YFP-treated IFNβ for the indicated periods. The amounts of precipitated DNA were determined by qPCR in duplicate using primers shown in A. The results represent one of four independent experiments, all of which gave similar results. The_right panel_ in D depicts H3.3-YFP incorporation in early time points. F, the ratio of H3.3-YFP and H3.1-YFP at the indicated times following IFNβ treatment.
FIGURE 2.
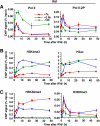
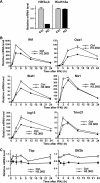
H3.3-YFP distribution in multiple IFN-inducible genes and in constitutively expressed genes. A–C, ChIP analysis was performed with anti-GFP antibody for the indicated IFN-inducible genes in H3.3-YFP-expressing NIH3T3 cells treated with IFNβ for 1, 3, 6, 12, 24, and 48 h as in Fig. 1. The gene map and the location of primers are shown underneath. D and_E_, ChIP analysis was performed for Gtf2b and Tbp, encoding TFIIB and TBP as above.
FIGURE 3.
Recruitment of RNA pol II and histone modification patterns on the_IfIt1_ gene following IFNβ treatment. ChIP analysis was performed for the Ifit1 gene using antibodies for total RNA pol II, pol II phosphorylated at serine 2 of the C-terminal domain (Pol II-2P), H3K4me3, diacetylated H3 (Lys-9 and Lys-14), H3K36me3, or H3K9me3 in NIH3T3 cells following IFNβ treatment for 1,3,6,12, 24, and 48 h, as in Fig. 1.
FIGURE 4.
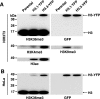
Global association of H3.3 with H3K36me3. Acid extracted histones from NIH3T3 (A) or HeLa (B) cells and those expressing H3.1-YFP and H3.3-YFP were subjected to immunoblot analysis using the antibodies specific for GFP or the indicated H3 modifications. See a similar association in IFN-treated cells in supplemental Fig. S4.
FIGURE 5.
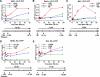
Reduction of H3.3 deposition following cell division. A, confluent NIH3T3 cells expressing H3.3-YFP were treated with IFNβ for 24 h. Cells were allowed to stand or were split into four plates and cultured for two additional days. Total cell yields on days 0, 1, and 3 are shown.B, transcript levels at the indicated times (left panel) and H3.3-YFP deposition in the 3′-coding region of indicated genes (right panel). See data for H3K36me in supplemental Fig. S5.
FIGURE 6.
Inhibition of IFN-stimulated gene expression in H3.3 knockdown (KD) cells. A, NIH3T3 cells were transduced with a vector for H3.3 shRNA or control (Ctrl) shRNA for 4 days, and levels of H3f3a,b (H3.3) or Hist1h3a (H3.1) transcripts were quantified by qRT-PCR. Values represent the average of three determinations ± S.D. B, the above cells were treated with IFNβ for 24 h, and the levels of indicated IFN-responsive transcripts were measured at the indicated times by qRT-PCR. C, levels of Tbp and_Gtf2b_ transcripts are measured in the above cells. Note that the_y_ axis is enlarged to emphasize the slight reduction in transcript expression in H3.3 knockdown cells.
Similar articles
- Histone H3.3 deposition at E2F-regulated genes is linked to transcription.
Daury L, Chailleux C, Bonvallet J, Trouche D. Daury L, et al. EMBO Rep. 2006 Jan;7(1):66-71. doi: 10.1038/sj.embor.7400561. EMBO Rep. 2006. PMID: 16258499 Free PMC article. - Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division.
Chow CM, Georgiou A, Szutorisz H, Maia e Silva A, Pombo A, Barahona I, Dargelos E, Canzonetta C, Dillon N. Chow CM, et al. EMBO Rep. 2005 Apr;6(4):354-60. doi: 10.1038/sj.embor.7400366. EMBO Rep. 2005. PMID: 15776021 Free PMC article. - Transcriptional activation triggers deposition and removal of the histone variant H3.3.
Schwartz BE, Ahmad K. Schwartz BE, et al. Genes Dev. 2005 Apr 1;19(7):804-14. doi: 10.1101/gad.1259805. Epub 2005 Mar 17. Genes Dev. 2005. PMID: 15774717 Free PMC article. - H3.3 turnover: a mechanism to poise chromatin for transcription, or a response to open chromatin?
Huang C, Zhu B. Huang C, et al. Bioessays. 2014 Jun;36(6):579-84. doi: 10.1002/bies.201400005. Epub 2014 Apr 3. Bioessays. 2014. PMID: 24700556 Review. - In and out: histone variant exchange in chromatin.
Jin J, Cai Y, Li B, Conaway RC, Workman JL, Conaway JW, Kusch T. Jin J, et al. Trends Biochem Sci. 2005 Dec;30(12):680-7. doi: 10.1016/j.tibs.2005.10.003. Epub 2005 Oct 28. Trends Biochem Sci. 2005. PMID: 16257529 Review.
Cited by
- Plasticity of histone modifications around Cidea and Cidec genes with secondary bile in the amelioration of developmentally-programmed hepatic steatosis.
Urmi JF, Itoh H, Muramatsu-Kato K, Kohmura-Kobayashi Y, Hariya N, Jain D, Tamura N, Uchida T, Suzuki K, Ogawa Y, Shiraki N, Mochizuki K, Kubota T, Kanayama N. Urmi JF, et al. Sci Rep. 2019 Nov 19;9(1):17100. doi: 10.1038/s41598-019-52943-7. Sci Rep. 2019. PMID: 31745102 Free PMC article. - Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation.
Martire S, Gogate AA, Whitmill A, Tafessu A, Nguyen J, Teng YC, Tastemel M, Banaszynski LA. Martire S, et al. Nat Genet. 2019 Jun;51(6):941-946. doi: 10.1038/s41588-019-0428-5. Epub 2019 May 31. Nat Genet. 2019. PMID: 31152160 Free PMC article. - Arabidopsis replacement histone variant H3.3 occupies promoters of regulated genes.
Shu H, Nakamura M, Siretskiy A, Borghi L, Moraes I, Wildhaber T, Gruissem W, Hennig L. Shu H, et al. Genome Biol. 2014 Mar 21;15(4):R62. doi: 10.1186/gb-2014-15-4-r62. Genome Biol. 2014. PMID: 24708891 Free PMC article. - Clinical immunity to malaria involves epigenetic reprogramming of innate immune cells.
Nideffer J, Ty M, Donato M, John R, Kajubi R, Ji X, Nankya F, Musinguzi K, Press KD, Yang N, Camanag K, Greenhouse B, Kamya M, Feeney ME, Dorsey G, Utz PJ, Pulendran B, Khatri P, Jagannathan P. Nideffer J, et al. PNAS Nexus. 2024 Aug 6;3(8):pgae325. doi: 10.1093/pnasnexus/pgae325. eCollection 2024 Aug. PNAS Nexus. 2024. PMID: 39161730 Free PMC article. - Molecular turnover, the H3.3 dilemma and organismal aging (hypothesis).
Saade E, Pirozhkova I, Aimbetov R, Lipinski M, Ogryzko V. Saade E, et al. Aging Cell. 2015 Jun;14(3):322-33. doi: 10.1111/acel.12332. Epub 2015 Feb 26. Aging Cell. 2015. PMID: 25720734 Free PMC article. Review.
References
- Henikoff, S., and Ahmad, K. (2005) Annu. Rev. Cell Dev. Biol. 21 133-153 - PubMed
- Sarma, K., and Reinberg, D. (2005) Nat. Rev. Mol. Cell Biol. 6 139-149 - PubMed
- Henikoff, S. (2008) Nat. Rev. Genet. 9 15-26 - PubMed
- Tagami, H., Ray-Gallet, D., Almouzni, G., and Nakatani, Y. (2004) Cell 116 51-61 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources