The interaction of the gammaherpesvirus 68 orf73 protein with cellular BET proteins affects the activation of cell cycle promoters - PubMed (original) (raw)
The interaction of the gammaherpesvirus 68 orf73 protein with cellular BET proteins affects the activation of cell cycle promoters
Matthias Ottinger et al. J Virol. 2009 May.
Abstract
Infection of mice with murine gammaherpesvirus 68 (MHV-68) provides a valuable animal model for gamma-2 herpesvirus (rhadinovirus) infection and pathogenesis. The MHV-68 orf73 protein has been shown to be required for the establishment of viral latency in vivo. This study describes a novel transcriptional activation function of the MHV-68 orf73 protein and identifies the cellular bromodomain containing BET proteins Brd2/RING3, Brd3/ORFX, and BRD4 as interaction partners for the MHV-68 orf73 protein. BET protein members are known to interact with acetylated histones, and Brd2 and Brd4 have been implicated in fundamental cellular processes, including cell cycle regulation and transcriptional regulation. Using MHV-68 orf73 peptide array assays, we identified Brd2 and Brd4 interaction sites in the orf73 protein. Mutation of one binding site led to a loss of the interaction with Brd2/4 but not the retinoblastoma protein Rb, to impaired chromatin association, and to a decreased ability to activate the BET-responsive cyclin D1, D2, and E promoters. The results therefore pinpoint the binding site for Brd2/4 in a rhadinoviral orf73 protein and suggest that the recruitment of a member of the BET protein family allows the MHV-68 orf73 protein to activate the promoters of G(1)/S cyclins. These findings point to parallels between the transcriptional activator functions of rhadinoviral orf73 proteins and papillomavirus E2 proteins.
Figures
FIG. 1.
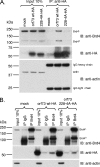
The MHV-68 orf73 protein interacts with the carboxy termini of the cellular BET proteins Brd2/RING3, Brd4S, and Brd3/ORFX. (A) Schematic depiction of the human BET proteins. BR1, bromodomain 1; BR2, bromodomain 2. (B) High degree of sequence conservation among human and murine ET domains. The human BET proteins are encoded by four genes, Brd2, Brd3, Brd4, and Brd6, and are characterized by two bromodomains and an ET domain. The BET protein sizes range from 722 to 1,362 aa for Brd4S and Brd4L, respectively. (B) ClustalW protein sequence alignment of human (hu) and murine (mu) BET protein 64-aa-long ET domains. ET domains of hu Brd2 (aa 640 to 703), mu Brd2 (aa 638 to 701), hu Brd3 (aa 570 to 633), mu Brd3 (aa 571 to 634), hu Brd4 (aa 608 to 671), mu Brd4 (aa 609 to 672), hu Brd6 (aa 508 to 571), and mu Brd6 (aa 504 to 567) were aligned. Boxed residues differ from the consensus sequence. The alignment shows 100% identity of the human and murine orthologous Brd2 ET domains, of human and murine Brd3 ET domains, and of human and murine Brd4 ET domains. The Brd6 ET domains of mice and humans are 95.3% identical. ET domain identity between paralogous BET proteins is also high, ranging from 84.4% to 92.2%. (C) Schematic of the GST-BET fusion proteins. Labels A, B, C, and D correspond to lane labeling in panel D. (D) The full-length MHV-68 orf73-HA protein interacted with GST-BET fusion proteins containing the ET domains of Brd2/RING3, Brd4, and Brd3/ORFX in GST pull-down assays. Bacterially expressed GST fusion proteins were bound to glutathione Sepharose resin and subsequently incubated with lysates from 293T cells transfected with MHV-68 orf73-HA expression plasmid. Ten percent of the input lysates was loaded for comparison. Top, anti-HA immunoblot (IB) used to detect the orf73 protein; bottom, anti-GST immunoblot used to verify expression of the GST-BET fusion proteins.
FIG. 2.
A peptide array assay identifies a binding site for purified Brd2 in the MHV-68 orf73 protein. (A) Schematic depiction of the baculovirus-expressed Brd2/RING3 protein that was purified and used for studies of direct interaction by peptide array assays, with results shown in panel D. BR1, bromodomain 1; BR2, bromodomain 2. (B) The purity of Brd2/RING3 was determined by SDS-PAGE followed by Coomassie staining. The identity of Brd2/RING2 was confirmed by anti-RING3 immunoblotting (IB). (C) Peptides representing the complete MHV-68 orf73 protein with each peptide overlapping with the previous one in 12 out of 15 residues were first incubated with SF9 lysate from untransfected SF9 insect cells and then probed with a primary rabbit anti-RING3 polyclonal antibody followed by incubation with a secondary AP-conjugated antibody to determine background staining. The nomenclature of peptides starts with 1A in the top left corner. (D) After the membrane was stripped, the same peptide array was incubated with affinity-purified Brd2/RING3 protein (see legend for panel B). Bound protein was detected as described for panel C. The peptide with the strongest signal intensity is marked with an asterisk. (E) Quantification of signal intensities shown in panel D minus background intensities shown in panel C, determined using Phoretix array 1.0 quantification software. (F) Sequences of the four peptides, 3W, 3X, 3Y, and 4A, that bound RING3, all containing the amino acids QAKKLK, corresponding to positions 226 to 231 in the MHV-68 orf73 protein.
FIG. 3.
A peptide array assay identifies a binding site for purified Brd4S in the MHV-68 orf73 protein. (A) Schematic depiction of the baculovirus-expressed Brd4S protein that was purified and used for studies of direct interaction by peptide array assays, with results shown in panel D. BR1, bromodomain 1; BR2, bromodomain 2. (B) The purity of Brd4S was determined by SDS-PAGE followed by Coomassie staining. The identity of Brd4S was confirmed by anti-His immunoblotting (IB). (C) A newly synthesized MHV-68 orf73 peptide array was first incubated with an anti-myc antibody followed by a secondary AP-conjugated antibody to determine background staining. (D) Next, the array was incubated with purified full-length Brd4S protein (see legend for panel B). Bound Brd4 protein was detected via its myc epitope tag. (E) Quantification of signal intensities shown in panel D minus background intensities shown in panel C. (F) Schematic depiction of the peptides that bound Brd4 (1D to 1G and 3X, 3Y, 4A, and 4B). (G) Schematic depiction of Brd2 and Brd4 binding sites in the MHV-68 orf73 protein, as determined by peptide array assays.
FIG. 4.
Interaction of MHV-68 orf73 protein mutants with Brd2, Brd4, and Rb. (A) Schematic depiction of MHV-68 orf73 mutants generated and used in this study. Labels A through J correspond to lane labeling in panels B, C, and D. (B) Co-IP of orf73 wt, orf73 mutants, and EGFP-Brd2. HA-tagged orf73 proteins were immunoprecipitated from lysates of 293T cells transfected with expression vectors for orf73 proteins and EGFP-Brd2 using an antibody to the HA tag. Immunoprecipitated proteins were analyzed by immunoblotting (IB) using an antibody to GFP. (C) Co-IP of orf73 wt, orf73 mutants, and EGFP-Brd4S. Experiments were performed as described for panel B. (D) Co-IP of orf73 wt, orf73 mutants, and cotransfected, myc-tagged Rb protein. Transfected, c-myc-tagged Rb was immunoprecipitated with an antibody to the c-myc epitope, and immunoprecipitates were analyzed by immunoblotting using an antibody to the HA epitope.
FIG. 5.
The MHV-68 orf73 protein forms complexes with endogenous Brd4 in B cells. BJAB cells transiently expressing the HA-tagged MHV-68 orf73 or orf73 228-4A protein were used for Co-IP experiments using anti-HA resin (A) and anti-Brd4 resin (B). Immunoblotting (IB) was performed using rabbit polyclonal anti-Brd4 serum (top panels), rat monoclonal anti-HA antibody (middle panels), and mouse monoclonal antiactin antibody (bottom panels). Ten percent of the lysates used for IP were loaded as input. (A) Co-IP using anti-HA resin reveals binding of Brd4L and Brd4S to MHV-68 orf73 wt but not the orf73 228-4A mutant. (B) Reciprocal Co-IP using anti-Brd4 resin. Normal IgG served as a negative control. With longer exposure, Brd4S becomes visible in the input lanes, as seen in panel A.
FIG. 6.
Decreased chromatin association of MHV-68 orf73 protein mutants. (A) Nuclear extracts of 293T cells with ectopically expressed MHV-68 orf73 mutants were subjected to increasing KCl concentrations to extract proteins. The supernatants of washes with 200 mM and 300 mM were analyzed by Western blotting. Expression levels of individual mutants were controlled in detergent lysates of transfected cells prior to washes with increasing KCl concentrations. IB, immunoblotting. (B) Summary of binding of orf73 wt and mutant proteins to Brd2, Brd4S, and pRB (results shown in Fig. 4), as well as extractability in high KCl concentrations. Labels A through J correspond to lane labeling in panel A. +, weak; ++, moderate; +++, strong; −, no interaction; n.a, not applicable.
FIG. 7.
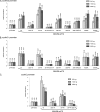
The MHV-68 orf73 protein activates promoters of cell cycle regulatory genes, and the BET interaction site KKLK (aa 228 to 231) in the orf73 protein is critical for this function. Transient luciferase reporter assays were performed with murine 3T3 fibroblasts. Cells were cotransfected with 50 ng of promoter luciferase reporter plasmids together with 500 ng, 1,000 ng, or 2,500 ng of empty vector (pVR1255) or different MHV-68 orf73 constructs (Fig. 4A). Relative luciferase activities compared to that for the empty vector were calculated, and mean values ± standard deviations from two representative experiments in duplicate are depicted. (A) Murine cyclin D2 promoter; (B) murine cyclin D1 promoter; (C) human cyclin E promoter.
Similar articles
- A structural basis for BRD2/4-mediated host chromatin interaction and oligomer assembly of Kaposi sarcoma-associated herpesvirus and murine gammaherpesvirus LANA proteins.
Hellert J, Weidner-Glunde M, Krausze J, Richter U, Adler H, Fedorov R, Pietrek M, Rückert J, Ritter C, Schulz TF, Lührs T. Hellert J, et al. PLoS Pathog. 2013;9(10):e1003640. doi: 10.1371/journal.ppat.1003640. Epub 2013 Oct 17. PLoS Pathog. 2013. PMID: 24146614 Free PMC article. - Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest.
Ottinger M, Christalla T, Nathan K, Brinkmann MM, Viejo-Borbolla A, Schulz TF. Ottinger M, et al. J Virol. 2006 Nov;80(21):10772-86. doi: 10.1128/JVI.00804-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928766 Free PMC article. - Brd/BET Proteins Influence the Genome-Wide Localization of the Kaposi's Sarcoma-Associated Herpesvirus and Murine Gammaherpesvirus Major Latency Proteins.
Lotke R, Schneeweiß U, Pietrek M, Günther T, Grundhoff A, Weidner-Glunde M, Schulz TF. Lotke R, et al. Front Microbiol. 2020 Oct 22;11:591778. doi: 10.3389/fmicb.2020.591778. eCollection 2020. Front Microbiol. 2020. PMID: 33193257 Free PMC article. - The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation.
Wu SY, Chiang CM. Wu SY, et al. J Biol Chem. 2007 May 4;282(18):13141-5. doi: 10.1074/jbc.R700001200. Epub 2007 Feb 28. J Biol Chem. 2007. PMID: 17329240 Review. - The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins.
Taniguchi Y. Taniguchi Y. Int J Mol Sci. 2016 Nov 7;17(11):1849. doi: 10.3390/ijms17111849. Int J Mol Sci. 2016. PMID: 27827996 Free PMC article. Review.
Cited by
- The latency-associated nuclear antigen, a multifunctional protein central to Kaposi's sarcoma-associated herpesvirus latency.
Ballestas ME, Kaye KM. Ballestas ME, et al. Future Microbiol. 2011 Dec;6(12):1399-413. doi: 10.2217/fmb.11.137. Future Microbiol. 2011. PMID: 22122438 Free PMC article. Review. - Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration.
Gupta SS, Maetzig T, Maertens GN, Sharif A, Rothe M, Weidner-Glunde M, Galla M, Schambach A, Cherepanov P, Schulz TF. Gupta SS, et al. J Virol. 2013 Dec;87(23):12721-36. doi: 10.1128/JVI.01942-13. Epub 2013 Sep 18. J Virol. 2013. PMID: 24049186 Free PMC article. - Murine Gammaherpesvirus 68 LANA and SOX Homologs Counteract ATM-Driven p53 Activity during Lytic Viral Replication.
Sifford JM, Stahl JA, Salinas E, Forrest JC. Sifford JM, et al. J Virol. 2015 Dec 16;90(5):2571-85. doi: 10.1128/JVI.02867-15. J Virol. 2015. PMID: 26676792 Free PMC article. - The Kaposi Sarcoma Herpesvirus Latency-associated Nuclear Antigen DNA Binding Domain Dorsal Positive Electrostatic Patch Facilitates DNA Replication and Episome Persistence.
Li S, Tan M, Juillard F, Ponnusamy R, Correia B, Simas JP, Carrondo MA, McVey CE, Kaye KM. Li S, et al. J Biol Chem. 2015 Nov 20;290(47):28084-28096. doi: 10.1074/jbc.M115.674622. Epub 2015 Sep 29. J Biol Chem. 2015. PMID: 26420481 Free PMC article. - Relevance of BET Family Proteins in SARS-CoV-2 Infection.
Lara-Ureña N, García-Domínguez M. Lara-Ureña N, et al. Biomolecules. 2021 Jul 30;11(8):1126. doi: 10.3390/biom11081126. Biomolecules. 2021. PMID: 34439792 Free PMC article. Review.
References
- Abbate, E. A., C. Voitenleitner, and M. R. Botchan. 2006. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol. Cell 24877-889. - PubMed
- An, F. Q., N. Compitello, E. Horwitz, M. Sramkoski, E. S. Knudsen, and R. Renne. 2005. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 2803862-3874. - PubMed
- Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284641-644. - PubMed
- Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311856-861. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous